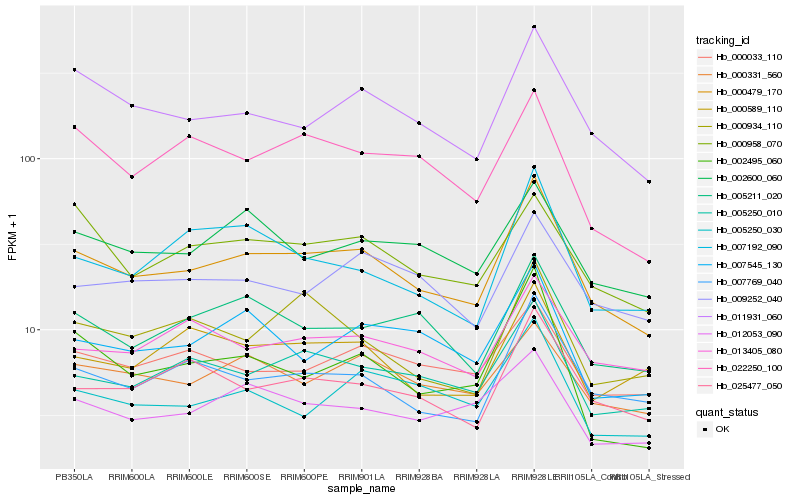
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_170 |
0.0 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 2 |
Hb_007769_040 |
0.0874654743 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 3 |
Hb_011931_060 |
0.0881740877 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 4 |
Hb_005250_010 |
0.0887905984 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 5 |
Hb_025477_050 |
0.0966548698 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_005211_020 |
0.1051431493 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_007545_130 |
0.1053261262 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005250_030 |
0.1060997497 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 9 |
Hb_000331_560 |
0.1062524054 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 10 |
Hb_007192_090 |
0.1063064192 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 11 |
Hb_012053_090 |
0.1075199781 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 12 |
Hb_013405_080 |
0.108961377 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 13 |
Hb_009252_040 |
0.1096709436 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000033_110 |
0.1098713491 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 15 |
Hb_002600_060 |
0.1115157236 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 16 |
Hb_002495_060 |
0.1129876663 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_022250_100 |
0.1135883384 |
- |
- |
calnexin, putative [Ricinus communis] |
| 18 |
Hb_000589_110 |
0.114049341 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000958_070 |
0.1163961512 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 20 |
Hb_000934_110 |
0.1166007856 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |