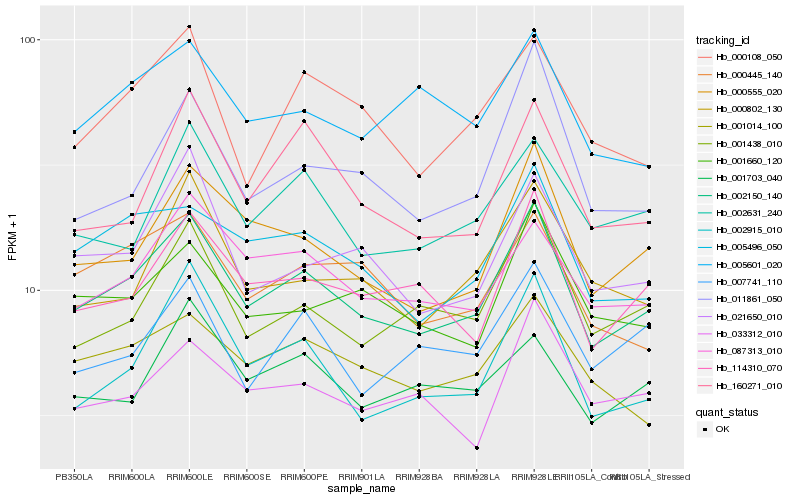
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_140 |
0.0 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 2 |
Hb_000555_020 |
0.0768037056 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_114310_070 |
0.082420695 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 4 |
Hb_005496_050 |
0.0839339271 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 5 |
Hb_087313_010 |
0.0857097419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001438_010 |
0.0883372011 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 7 |
Hb_021650_010 |
0.0917384343 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 8 |
Hb_002915_010 |
0.091863333 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 9 |
Hb_001660_120 |
0.0926442101 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001014_100 |
0.0929036864 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 11 |
Hb_000108_050 |
0.0929741685 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 12 |
Hb_002631_240 |
0.0932785367 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 13 |
Hb_000445_140 |
0.0955837975 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 14 |
Hb_011861_050 |
0.0961507435 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_000802_130 |
0.097316263 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 16 |
Hb_005601_020 |
0.0977572461 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_160271_010 |
0.098357784 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_033312_010 |
0.0993447411 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_007741_110 |
0.1004733627 |
- |
- |
- |
| 20 |
Hb_001703_040 |
0.101016919 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |