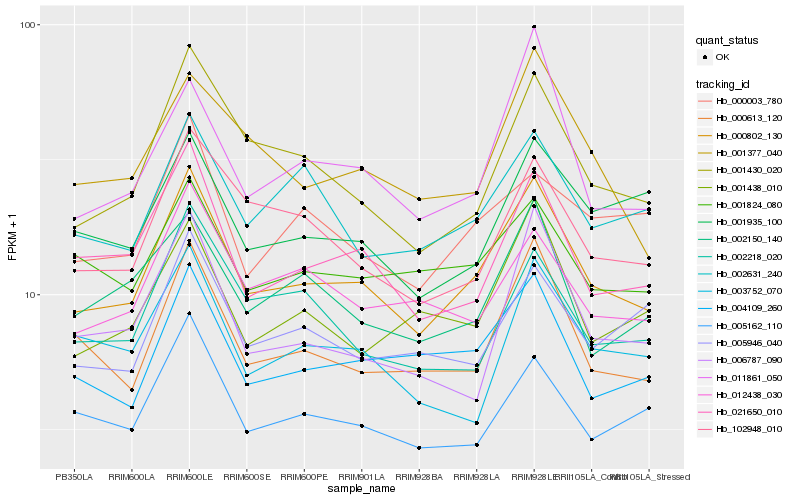
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000802_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 2 |
Hb_000613_120 |
0.0801213665 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_021650_010 |
0.0812296979 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 4 |
Hb_001430_020 |
0.0829062365 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_004109_260 |
0.0846147091 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 6 |
Hb_011861_050 |
0.0914284198 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_102948_010 |
0.0927624916 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012438_030 |
0.0951159759 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 9 |
Hb_005162_110 |
0.0952047547 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 10 |
Hb_001377_040 |
0.0955933873 |
- |
- |
chitinase, putative [Ricinus communis] |
| 11 |
Hb_001438_010 |
0.0959724184 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001824_080 |
0.096480017 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 13 |
Hb_002150_140 |
0.097316263 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 14 |
Hb_002631_240 |
0.0976191322 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 15 |
Hb_000003_780 |
0.1007685311 |
- |
- |
hexokinase [Manihot esculenta] |
| 16 |
Hb_002218_020 |
0.1023945638 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 17 |
Hb_001935_100 |
0.1026859235 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 18 |
Hb_006787_090 |
0.1029718723 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 19 |
Hb_003752_070 |
0.1047619131 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005946_040 |
0.1051457545 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |