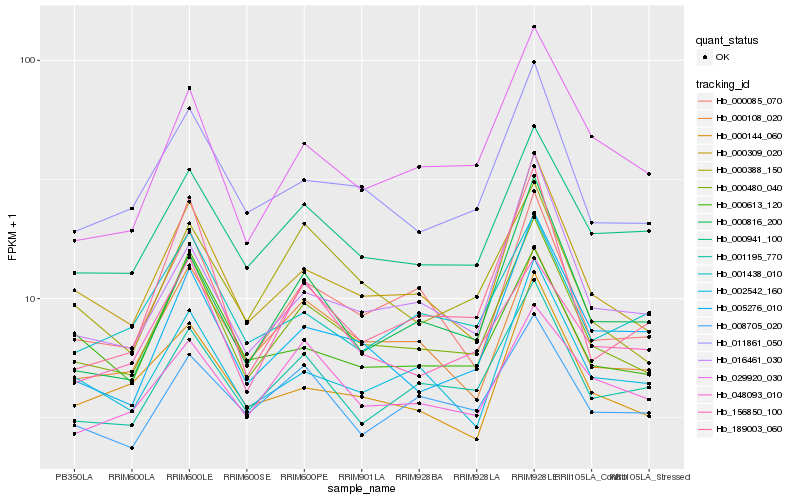
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000480_040 |
0.0 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 2 |
Hb_000309_020 |
0.0585804822 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 3 |
Hb_011861_050 |
0.0669674321 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_000085_070 |
0.070120588 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000941_100 |
0.0762723981 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 6 |
Hb_001195_770 |
0.0781303898 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 7 |
Hb_000108_020 |
0.0790616678 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 8 |
Hb_002542_160 |
0.086785664 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000816_200 |
0.0880298044 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 10 |
Hb_000388_150 |
0.0890213437 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 11 |
Hb_029920_030 |
0.0912194138 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_016461_030 |
0.0914606657 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_008705_020 |
0.0922609008 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_005276_010 |
0.0940084833 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 15 |
Hb_000144_060 |
0.0940721388 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000613_120 |
0.0943620609 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001438_010 |
0.0954008572 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 18 |
Hb_048093_010 |
0.0979339861 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_156850_100 |
0.0996630712 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 20 |
Hb_189003_060 |
0.0999248511 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |