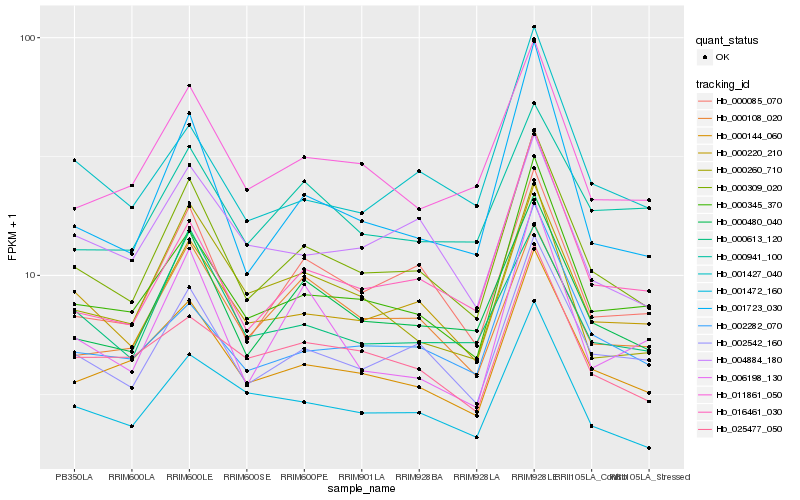
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000309_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 2 |
Hb_002542_160 |
0.0558405396 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000480_040 |
0.0585804822 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 4 |
Hb_000220_210 |
0.0636042017 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000345_370 |
0.0663271226 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 6 |
Hb_000085_070 |
0.0690120947 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000144_060 |
0.069063168 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001472_160 |
0.0808805234 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 9 |
Hb_011861_050 |
0.0819463521 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_016461_030 |
0.082031736 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001723_030 |
0.0850451481 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000613_120 |
0.0923133231 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_025477_050 |
0.0928374967 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_002282_070 |
0.0929289113 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 15 |
Hb_000108_020 |
0.0947698835 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 16 |
Hb_001427_040 |
0.0958747488 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 17 |
Hb_004884_180 |
0.0972903512 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000260_710 |
0.0973482325 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000941_100 |
0.1004260082 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 20 |
Hb_006198_130 |
0.1034115146 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |