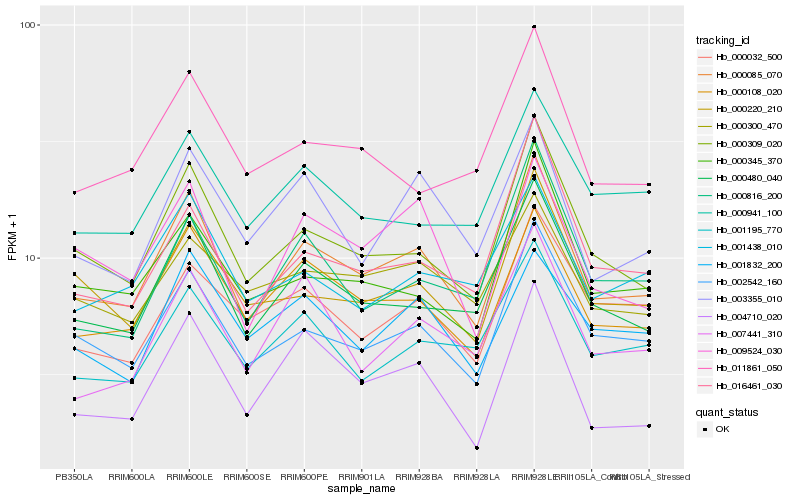
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_070 |
0.0 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000108_020 |
0.0687771377 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 3 |
Hb_000309_020 |
0.0690120947 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 4 |
Hb_000480_040 |
0.070120588 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 5 |
Hb_002542_160 |
0.071305257 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003355_010 |
0.0924625956 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 7 |
Hb_000220_210 |
0.0929097264 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 8 |
Hb_009524_030 |
0.0945118004 |
- |
- |
PREDICTED: uncharacterized protein LOC105650971 [Jatropha curcas] |
| 9 |
Hb_016461_030 |
0.0956936743 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_001195_770 |
0.0970592742 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 11 |
Hb_000345_370 |
0.0992898842 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 12 |
Hb_004710_020 |
0.09960086 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 13 |
Hb_007441_310 |
0.1008290394 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001832_200 |
0.1017735665 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001438_010 |
0.1021255783 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011861_050 |
0.10269963 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_000941_100 |
0.1030268174 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 18 |
Hb_000816_200 |
0.1030808849 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 19 |
Hb_000032_500 |
0.1036372994 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 20 |
Hb_000300_470 |
0.1037718797 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |