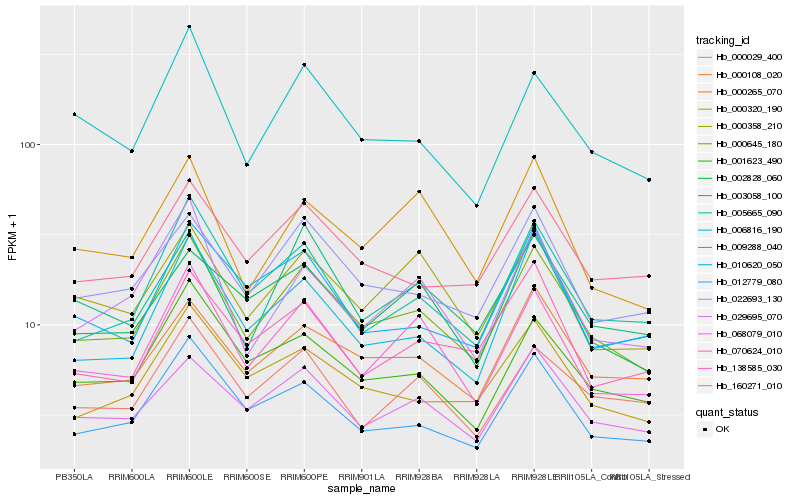
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000645_180 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 2 |
Hb_068079_010 |
0.0746748231 |
- |
- |
- |
| 3 |
Hb_022693_130 |
0.0762816658 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_012779_080 |
0.0765105139 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000108_020 |
0.0783307757 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 6 |
Hb_070624_010 |
0.0786866769 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 7 |
Hb_000320_190 |
0.0818969378 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_005665_090 |
0.0831025011 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 9 |
Hb_160271_010 |
0.0848646057 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001623_490 |
0.0873563724 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003058_100 |
0.0913477878 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002828_060 |
0.0930192242 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_006816_190 |
0.0945795468 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 14 |
Hb_000358_210 |
0.0955278142 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 15 |
Hb_000029_400 |
0.0972676895 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000265_070 |
0.0976712091 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 17 |
Hb_009288_040 |
0.098251031 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 18 |
Hb_010620_050 |
0.0994255619 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 19 |
Hb_138585_030 |
0.0999538645 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 20 |
Hb_029695_070 |
0.1003080348 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |