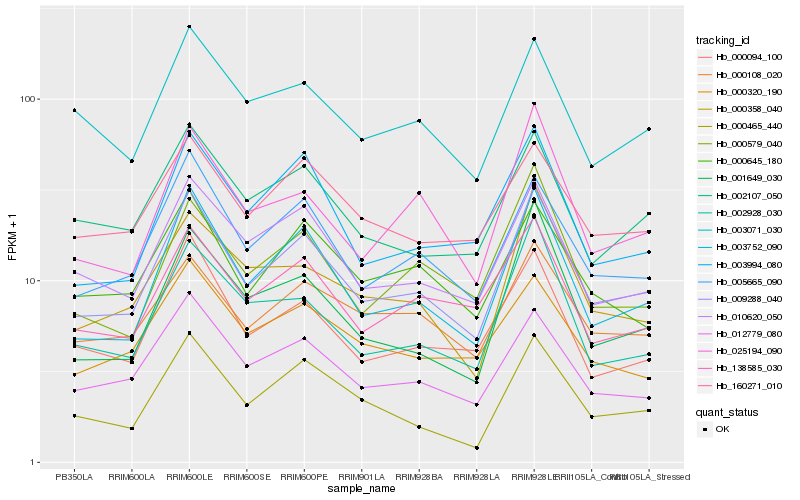
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009288_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 2 |
Hb_003752_090 |
0.0594450584 |
- |
- |
chitinase, putative [Ricinus communis] |
| 3 |
Hb_005665_090 |
0.0717073985 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 4 |
Hb_012779_080 |
0.0762527146 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 5 |
Hb_010620_050 |
0.0813169638 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 6 |
Hb_000094_100 |
0.0894580287 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 7 |
Hb_000465_440 |
0.0908014598 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 8 |
Hb_000108_020 |
0.0910258129 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 9 |
Hb_002107_050 |
0.0912133344 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 10 |
Hb_003994_080 |
0.0920536129 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 11 |
Hb_138585_030 |
0.0922508316 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 12 |
Hb_025194_090 |
0.0931366774 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 13 |
Hb_001649_030 |
0.0952959733 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003071_030 |
0.0967476167 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000358_040 |
0.0973065188 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002928_030 |
0.0974068579 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 17 |
Hb_160271_010 |
0.0979028763 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000645_180 |
0.098251031 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000579_040 |
0.0988365828 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 20 |
Hb_000320_190 |
0.1004131941 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |