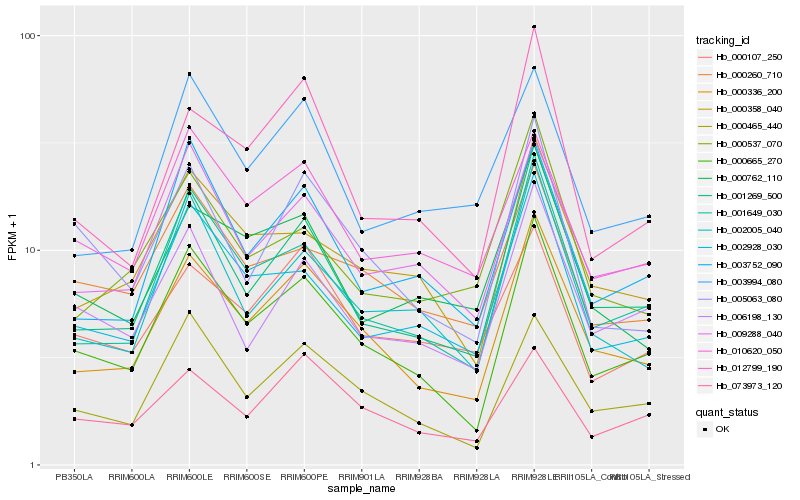
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_200 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_000665_270 |
0.0782761566 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 3 |
Hb_001269_500 |
0.07856038 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 4 |
Hb_001649_030 |
0.0930828129 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000465_440 |
0.0967971071 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 6 |
Hb_000358_040 |
0.1114564232 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009288_040 |
0.1129702392 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 8 |
Hb_073973_120 |
0.113841065 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_012799_190 |
0.1140128183 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 10 |
Hb_002005_040 |
0.117141278 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_003752_090 |
0.1191390303 |
- |
- |
chitinase, putative [Ricinus communis] |
| 12 |
Hb_002928_030 |
0.1242318586 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 13 |
Hb_000260_710 |
0.1242692813 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000762_110 |
0.1244179493 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003994_080 |
0.1257989579 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 16 |
Hb_006198_130 |
0.1267066772 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 17 |
Hb_000107_250 |
0.1279099423 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 18 |
Hb_000537_070 |
0.1293640998 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 19 |
Hb_010620_050 |
0.1301846332 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 20 |
Hb_005063_080 |
0.1302893225 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |