| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
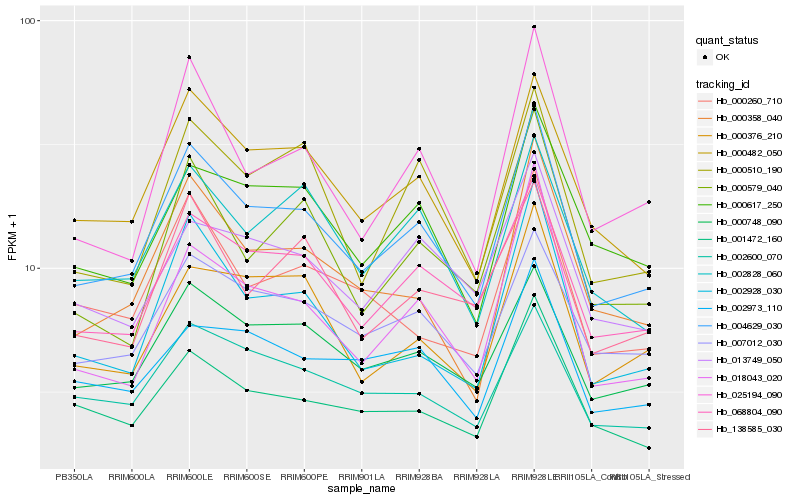
Hb_004629_030 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_000482_050 |
0.0652748175 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_068804_090 |
0.0679907006 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 4 |
Hb_000748_090 |
0.0764394383 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 5 |
Hb_007012_030 |
0.0790085325 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000358_040 |
0.0830280694 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000617_250 |
0.0849540438 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_025194_090 |
0.0859559635 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 9 |
Hb_002928_030 |
0.0860945634 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 10 |
Hb_002828_060 |
0.0874336828 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000376_210 |
0.0874369748 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 12 |
Hb_018043_020 |
0.0883701579 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000510_190 |
0.0906003412 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 14 |
Hb_002600_070 |
0.0909806123 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 15 |
Hb_001472_160 |
0.0920303735 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 16 |
Hb_013749_050 |
0.0925335163 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000579_040 |
0.0937481209 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 18 |
Hb_138585_030 |
0.0946245899 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 19 |
Hb_000260_710 |
0.0958547922 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002973_110 |
0.0972857822 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |