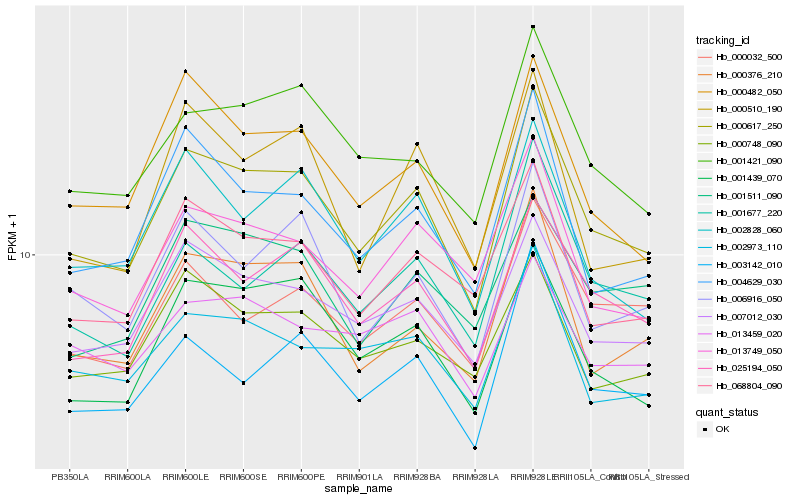
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_250 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007012_030 |
0.072320648 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_025194_050 |
0.0743932032 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001421_090 |
0.0809841934 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 5 |
Hb_000376_210 |
0.0829394944 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 6 |
Hb_000482_050 |
0.0838437288 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_068804_090 |
0.0839230143 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 8 |
Hb_004629_030 |
0.0849540438 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_001439_070 |
0.0853771154 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_013749_050 |
0.0855247393 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006916_050 |
0.085678821 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_002828_060 |
0.0895612489 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002973_110 |
0.0897198777 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_001677_220 |
0.0934491735 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 15 |
Hb_000748_090 |
0.0938038927 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 16 |
Hb_000032_500 |
0.0951042492 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 17 |
Hb_003142_010 |
0.0965829312 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 18 |
Hb_001511_090 |
0.0987284461 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 19 |
Hb_000510_190 |
0.1009299444 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 20 |
Hb_013459_020 |
0.1014244599 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |