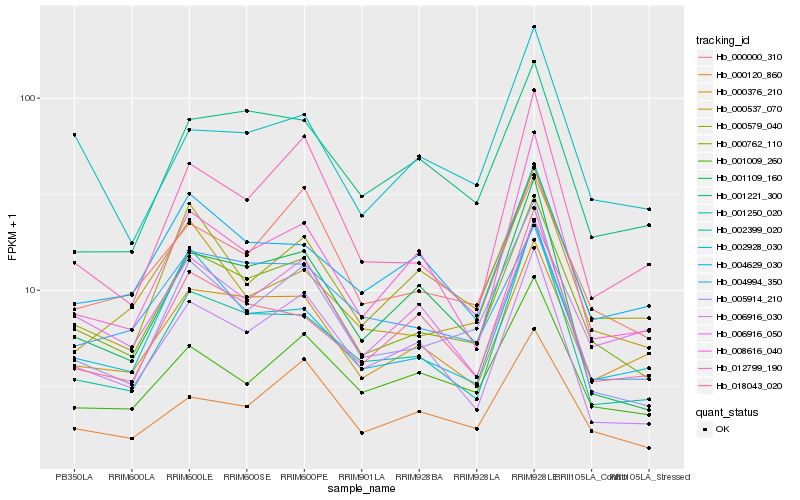
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000762_110 |
0.0 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_005914_210 |
0.0875832219 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 3 |
Hb_000376_210 |
0.091063631 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 4 |
Hb_001250_020 |
0.0964293423 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002928_030 |
0.1003346227 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 6 |
Hb_000120_860 |
0.1014891287 |
- |
- |
nucellin, putative [Ricinus communis] |
| 7 |
Hb_000579_040 |
0.1030844482 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 8 |
Hb_006916_030 |
0.1032780541 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 9 |
Hb_001009_260 |
0.1042860674 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000000_310 |
0.1046417499 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_001109_160 |
0.1082820135 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_018043_020 |
0.1120254391 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004629_030 |
0.1126091396 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_006916_050 |
0.1138585538 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 15 |
Hb_002399_020 |
0.1139495314 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 16 |
Hb_008616_040 |
0.1141655183 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000537_070 |
0.116378531 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 18 |
Hb_004994_350 |
0.1164065407 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001221_300 |
0.116483378 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_012799_190 |
0.1167644693 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |