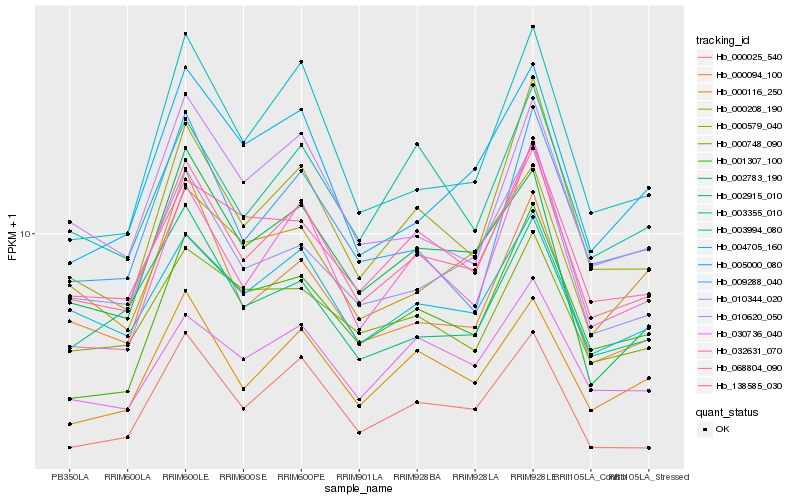
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138585_030 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 2 |
Hb_004705_160 |
0.0698495399 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 3 |
Hb_010344_020 |
0.0753677824 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000116_250 |
0.0761457922 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 5 |
Hb_000579_040 |
0.0789175694 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 6 |
Hb_010620_050 |
0.0802519517 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 7 |
Hb_068804_090 |
0.0832520874 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 8 |
Hb_030736_040 |
0.0842430146 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003994_080 |
0.0850962441 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 10 |
Hb_003355_010 |
0.0865576062 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 11 |
Hb_032631_070 |
0.0865701383 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_002783_190 |
0.0886426486 |
- |
- |
PREDICTED: uncharacterized protein LOC105635339 [Jatropha curcas] |
| 13 |
Hb_001307_100 |
0.0889787928 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_000094_100 |
0.0890693872 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 15 |
Hb_000748_090 |
0.0893509651 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 16 |
Hb_000025_540 |
0.0897390995 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 17 |
Hb_000208_190 |
0.0899832308 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 18 |
Hb_005000_080 |
0.0909676122 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 19 |
Hb_002915_010 |
0.0913082316 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 20 |
Hb_009288_040 |
0.0922508316 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |