| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
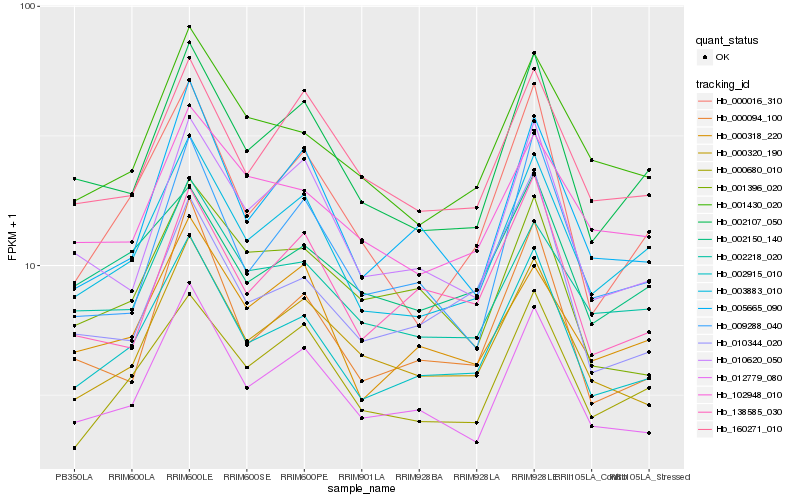
Hb_002915_010 |
0.0 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 2 |
Hb_012779_080 |
0.0652644832 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_003883_010 |
0.0697528606 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 4 |
Hb_000320_190 |
0.0768823518 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002107_050 |
0.0821734357 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 6 |
Hb_000094_100 |
0.0842169159 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 7 |
Hb_000680_010 |
0.0879524078 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 8 |
Hb_001430_020 |
0.0888089028 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_005665_090 |
0.0900963981 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 10 |
Hb_002218_020 |
0.0906550575 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 11 |
Hb_138585_030 |
0.0913082316 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 12 |
Hb_002150_140 |
0.091863333 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 13 |
Hb_010344_020 |
0.0935439921 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000016_310 |
0.0948142986 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_010620_050 |
0.0963462105 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 16 |
Hb_160271_010 |
0.0965356488 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000318_220 |
0.0975171997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009288_040 |
0.1015558296 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 19 |
Hb_102948_010 |
0.10193533 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001396_020 |
0.1027100129 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |