| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
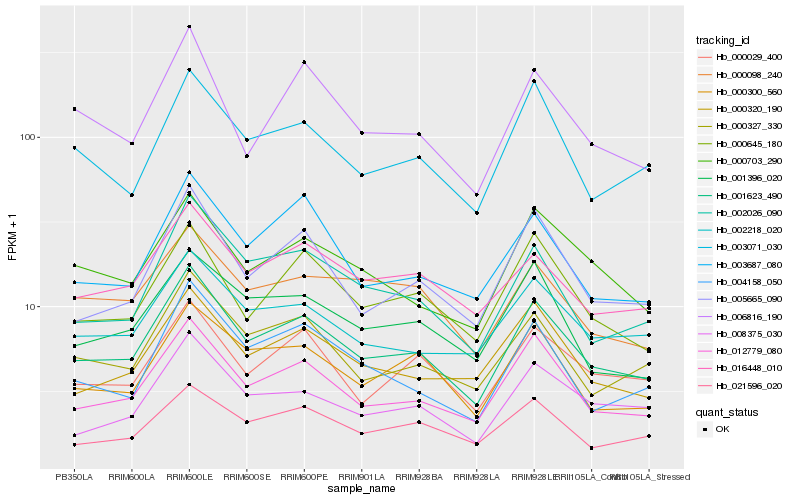
Hb_001623_490 |
0.0 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012779_080 |
0.0583468092 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000327_330 |
0.0791184319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005665_090 |
0.0792802075 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 5 |
Hb_002026_090 |
0.0805192589 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 6 |
Hb_002218_020 |
0.0859903806 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_016448_010 |
0.0868499824 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 8 |
Hb_000645_180 |
0.0873563724 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000300_560 |
0.0875768279 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003071_030 |
0.0876231304 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000320_190 |
0.0880693836 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_008375_030 |
0.0887927409 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 13 |
Hb_000029_400 |
0.0896971318 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003687_080 |
0.0897941574 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 15 |
Hb_000703_290 |
0.0905335745 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 16 |
Hb_001396_020 |
0.090868389 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 17 |
Hb_006816_190 |
0.0910344599 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 18 |
Hb_004158_050 |
0.0918483087 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 19 |
Hb_021596_020 |
0.092521595 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 20 |
Hb_000098_240 |
0.0970634824 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |