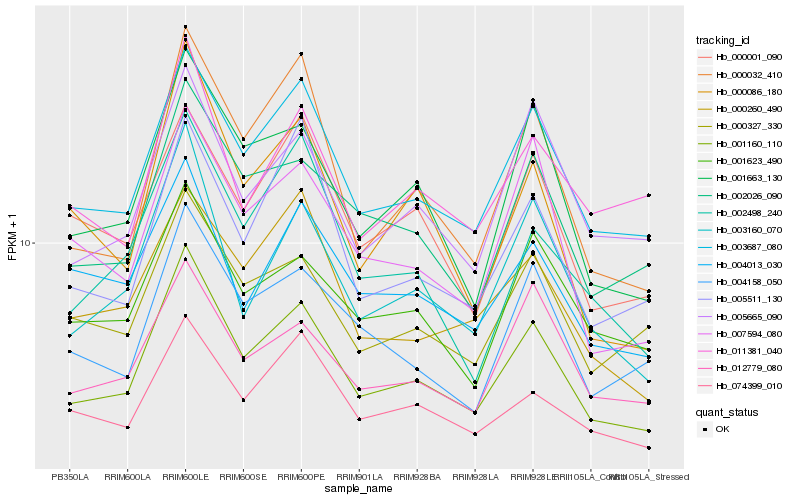
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_110 |
0.0 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 2 |
Hb_003687_080 |
0.0669290865 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 3 |
Hb_003160_070 |
0.0843552565 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 4 |
Hb_001663_130 |
0.084379538 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 5 |
Hb_002026_090 |
0.0900302738 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 6 |
Hb_000032_410 |
0.0933890806 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 7 |
Hb_004158_050 |
0.101134139 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 8 |
Hb_074399_010 |
0.1013803286 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 9 |
Hb_001623_490 |
0.1016580905 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000327_330 |
0.1021046618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005665_090 |
0.1022803919 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 12 |
Hb_007594_080 |
0.1040047412 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 13 |
Hb_000086_180 |
0.1056388217 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 14 |
Hb_012779_080 |
0.1069167414 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 15 |
Hb_005511_130 |
0.1076335587 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 16 |
Hb_004013_030 |
0.1085201208 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 17 |
Hb_000001_090 |
0.1087438913 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 18 |
Hb_000260_490 |
0.1098934233 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002498_240 |
0.111077018 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011381_040 |
0.113899955 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |