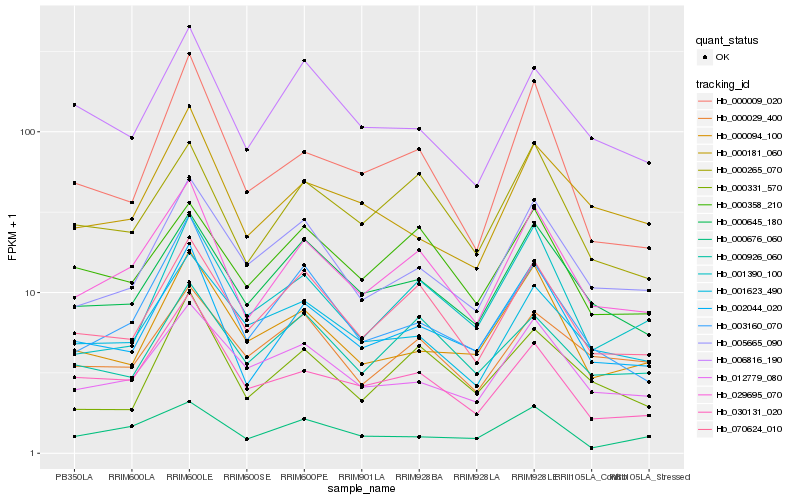
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029695_070 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002044_020 |
0.0805403696 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 3 |
Hb_070624_010 |
0.0908418308 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 4 |
Hb_000265_070 |
0.098546516 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 5 |
Hb_000645_180 |
0.1003080348 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 6 |
Hb_003160_070 |
0.1087888965 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 7 |
Hb_005665_090 |
0.1097011685 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 8 |
Hb_012779_080 |
0.1119146544 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_030131_020 |
0.1174444777 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 10 |
Hb_001623_490 |
0.1186723611 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000358_210 |
0.1217461325 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 12 |
Hb_001390_100 |
0.1220874528 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000331_570 |
0.1226876196 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 14 |
Hb_000926_060 |
0.1254194503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006816_190 |
0.1267605573 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 16 |
Hb_000181_060 |
0.1273316531 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000029_400 |
0.1273721578 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000009_020 |
0.1276981178 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000676_060 |
0.1277353573 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 20 |
Hb_000094_100 |
0.1280256993 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |