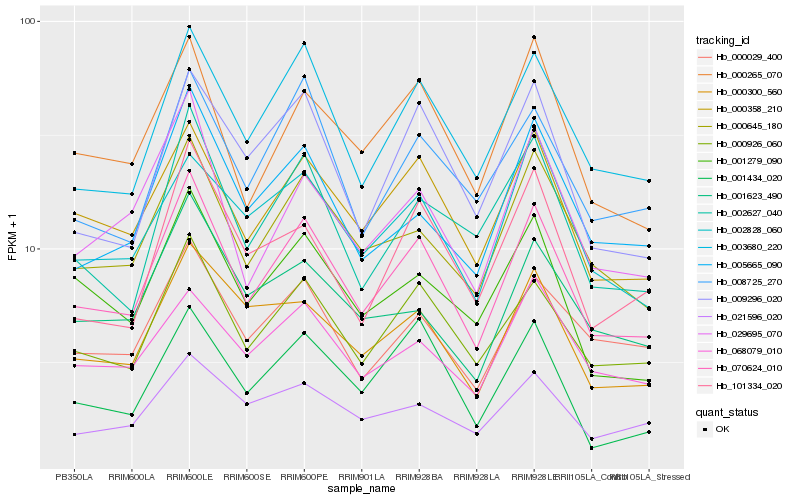
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070624_010 |
0.0 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 2 |
Hb_000926_060 |
0.0639389755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000358_210 |
0.0726131599 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 4 |
Hb_003680_220 |
0.0743131332 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 5 |
Hb_000645_180 |
0.0786866769 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 6 |
Hb_000265_070 |
0.0859625842 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 7 |
Hb_029695_070 |
0.0908418308 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001279_090 |
0.0931831135 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 9 |
Hb_000300_560 |
0.0935389744 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002627_040 |
0.0937126116 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_000029_400 |
0.0938050229 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009296_020 |
0.0950395206 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_005665_090 |
0.0957001026 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 14 |
Hb_008725_270 |
0.0986610794 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 15 |
Hb_001434_020 |
0.1017366313 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 16 |
Hb_101334_020 |
0.1018778884 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 17 |
Hb_001623_490 |
0.1048263064 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 18 |
Hb_021596_020 |
0.1056369643 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 19 |
Hb_002828_060 |
0.1060654554 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_068079_010 |
0.1063137757 |
- |
- |
- |