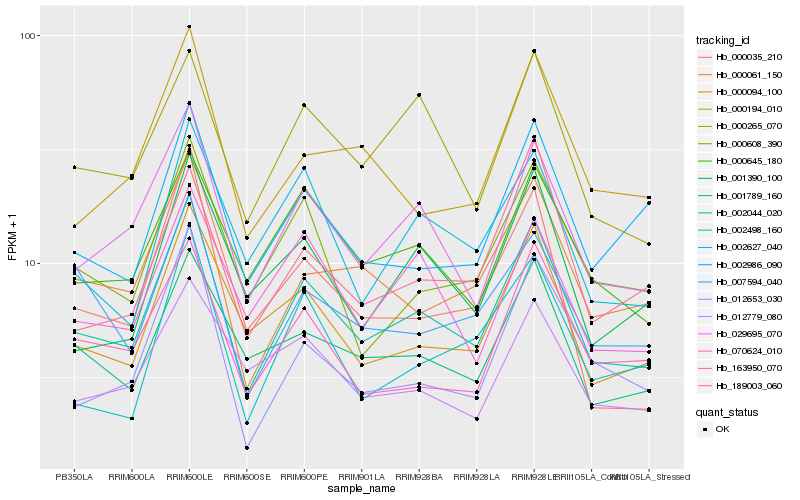
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002044_020 |
0.0 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 2 |
Hb_029695_070 |
0.0805403696 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000094_100 |
0.0965254036 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 4 |
Hb_001789_160 |
0.1054640026 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 5 |
Hb_000061_150 |
0.1059682947 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000645_180 |
0.1076203439 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_000265_070 |
0.1093495636 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 8 |
Hb_000608_390 |
0.1106702206 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |
| 9 |
Hb_163950_070 |
0.1119111456 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 10 |
Hb_012653_030 |
0.1146156035 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_001390_100 |
0.1147984125 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000194_010 |
0.1169331286 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002627_040 |
0.1173031194 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 14 |
Hb_002986_090 |
0.1186712388 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_070624_010 |
0.1192343245 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 16 |
Hb_007594_040 |
0.1192912375 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_189003_060 |
0.1197664723 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000035_210 |
0.1214188119 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 19 |
Hb_002498_160 |
0.1218645886 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 20 |
Hb_012779_080 |
0.1228788944 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |