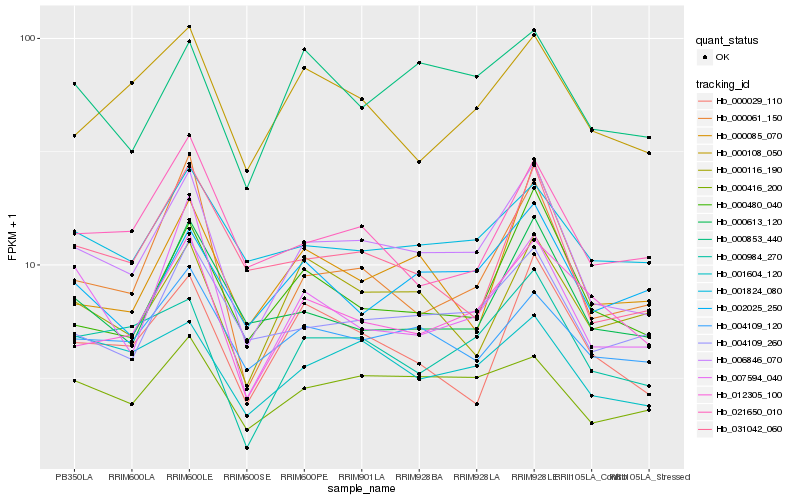
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_070 |
0.0 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_000984_270 |
0.1034151801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000416_200 |
0.1054165375 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 4 |
Hb_004109_260 |
0.1075782108 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 5 |
Hb_000061_150 |
0.108765903 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000480_040 |
0.1115317696 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 7 |
Hb_001604_120 |
0.1147616114 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004109_120 |
0.1152510017 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012305_100 |
0.1152847736 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000613_120 |
0.1176667442 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000029_110 |
0.1194187186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007594_040 |
0.1206823538 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000853_440 |
0.1209308522 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 14 |
Hb_001824_080 |
0.1211625195 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 15 |
Hb_031042_060 |
0.1211894612 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_000085_070 |
0.1220928818 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000108_050 |
0.1225016839 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 18 |
Hb_002025_250 |
0.122711489 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 19 |
Hb_000116_190 |
0.123304132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_021650_010 |
0.1251031036 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |