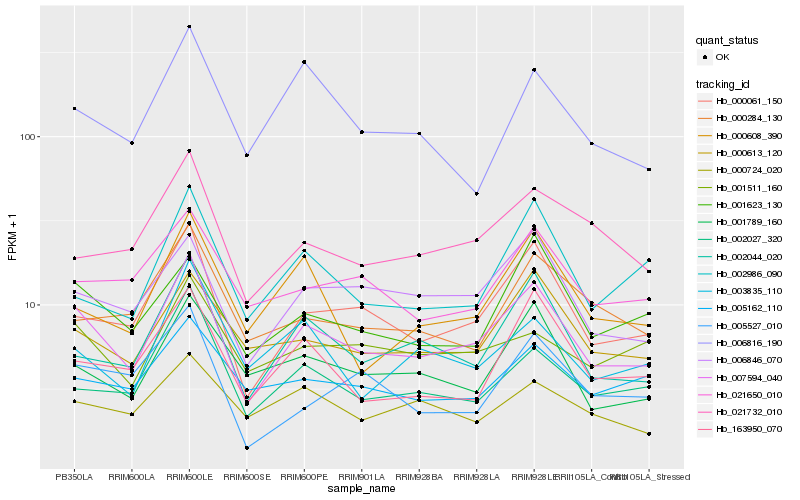
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007594_040 |
0.0 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000061_150 |
0.1011762482 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002044_020 |
0.1192912375 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 4 |
Hb_006846_070 |
0.1206823538 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_000613_120 |
0.1276319356 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001789_160 |
0.1309196413 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 7 |
Hb_002027_320 |
0.1340224938 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 8 |
Hb_163950_070 |
0.1364411433 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 9 |
Hb_001623_130 |
0.1379330536 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_021732_010 |
0.1379602252 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 11 |
Hb_021650_010 |
0.1400619135 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 12 |
Hb_001511_160 |
0.1417422315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003835_110 |
0.1430496933 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000608_390 |
0.1437782844 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |
| 15 |
Hb_005162_110 |
0.1443439746 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 16 |
Hb_005527_010 |
0.1452304985 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000284_130 |
0.1457169465 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 18 |
Hb_002986_090 |
0.1465071119 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_006816_190 |
0.1469250503 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 20 |
Hb_000724_020 |
0.1472093711 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |