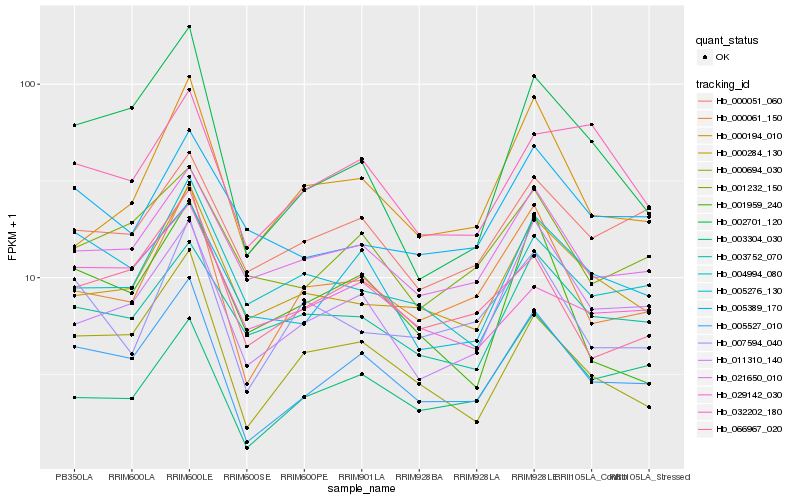
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005527_010 |
0.0 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000061_150 |
0.1086597551 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000694_030 |
0.1168385371 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 4 |
Hb_011310_140 |
0.1260913614 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 5 |
Hb_002701_120 |
0.1268283076 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 6 |
Hb_005276_130 |
0.1268561305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_066967_020 |
0.1276040505 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |
| 8 |
Hb_021650_010 |
0.1294101779 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 9 |
Hb_001232_150 |
0.1352367103 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 10 |
Hb_000284_130 |
0.1398141703 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 11 |
Hb_007594_040 |
0.1452304985 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_003304_030 |
0.1452748878 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000051_060 |
0.1473422023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003752_070 |
0.1493665832 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000194_010 |
0.1528418463 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_029142_030 |
0.1530588126 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 17 |
Hb_001959_240 |
0.1555408811 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_004994_080 |
0.1565894196 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_032202_180 |
0.1576093094 |
- |
- |
PREDICTED: protein-ribulosamine 3-kinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005389_170 |
0.157871568 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |