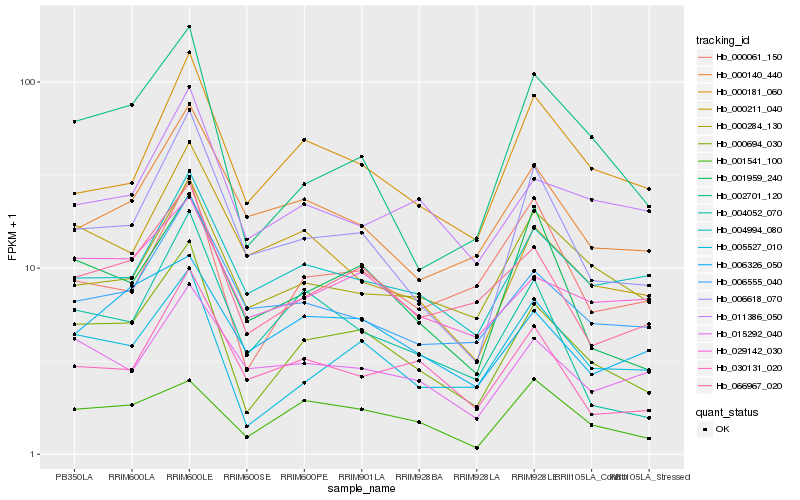
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000694_030 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 2 |
Hb_005527_010 |
0.1168385371 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_066967_020 |
0.1227331327 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |
| 4 |
Hb_006618_070 |
0.1277103949 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002701_120 |
0.1284386923 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 6 |
Hb_001959_240 |
0.1390251485 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_029142_030 |
0.1429628386 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 8 |
Hb_006555_040 |
0.148696678 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 9 |
Hb_004994_080 |
0.1508449419 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_030131_020 |
0.1520770516 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 11 |
Hb_000181_060 |
0.1521677524 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001541_100 |
0.1533333768 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 13 |
Hb_000211_040 |
0.1535231614 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 14 |
Hb_004052_070 |
0.1536947307 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 15 |
Hb_011386_050 |
0.1560868411 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000284_130 |
0.1576370831 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 17 |
Hb_000140_440 |
0.1595149473 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 18 |
Hb_000061_150 |
0.1597222019 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_015292_040 |
0.1600175568 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 20 |
Hb_006326_050 |
0.1601578906 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |