| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
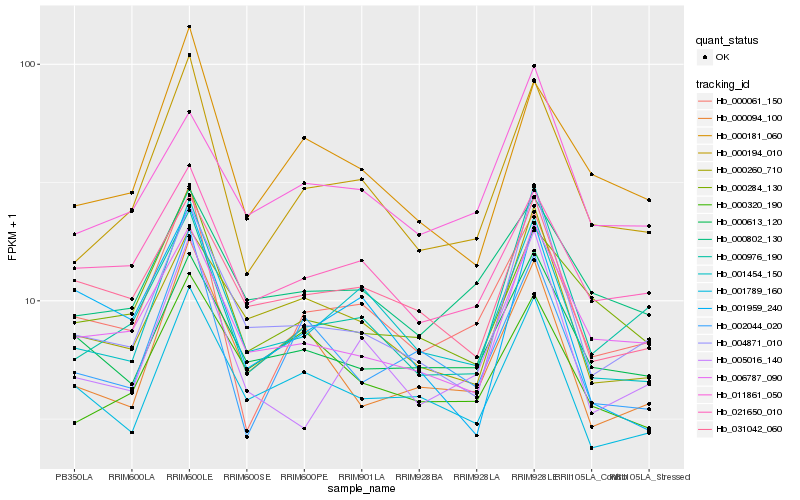
Hb_001454_150 |
0.0 |
- |
- |
isoleucyl tRNA synthetase, putative [Ricinus communis] |
| 2 |
Hb_000194_010 |
0.0897770333 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001789_160 |
0.0971200358 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 4 |
Hb_005016_140 |
0.1014766142 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_031042_060 |
0.1016646324 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000260_710 |
0.1098797917 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004871_010 |
0.1100746327 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 8 |
Hb_000061_150 |
0.1134090821 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_021650_010 |
0.1154399349 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 10 |
Hb_000613_120 |
0.1232151581 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000181_060 |
0.1246737476 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000802_130 |
0.1276309386 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 13 |
Hb_011861_050 |
0.1289887748 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_001959_240 |
0.1293553355 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_006787_090 |
0.1298089675 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 16 |
Hb_000094_100 |
0.13105142 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 17 |
Hb_000976_190 |
0.131497347 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 18 |
Hb_000320_190 |
0.1324797704 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000284_130 |
0.1329944058 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 20 |
Hb_002044_020 |
0.1349623419 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |