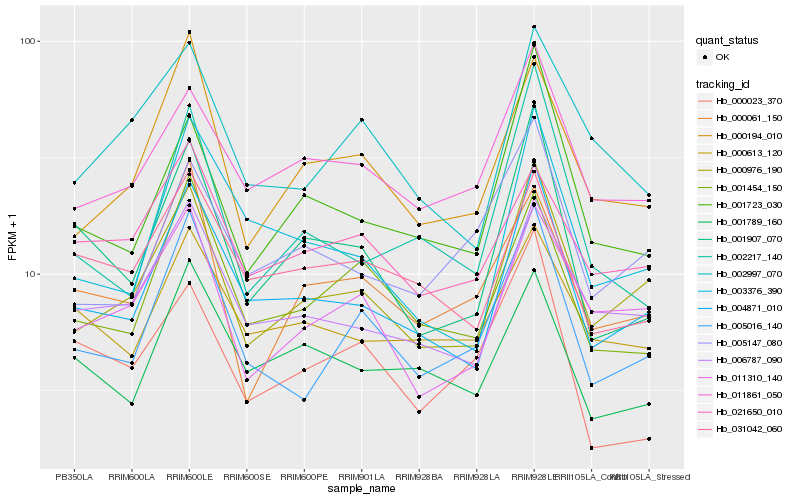
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005016_140 |
0.0 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001454_150 |
0.1014766142 |
- |
- |
isoleucyl tRNA synthetase, putative [Ricinus communis] |
| 3 |
Hb_004871_010 |
0.1163655765 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 4 |
Hb_000976_190 |
0.1192030939 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 5 |
Hb_000194_010 |
0.1280269796 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000061_150 |
0.1368107282 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011310_140 |
0.1414923263 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 8 |
Hb_006787_090 |
0.1437563842 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 9 |
Hb_001907_070 |
0.1453522184 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 10 |
Hb_005147_080 |
0.1463028372 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_011861_050 |
0.148068026 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_003376_390 |
0.1483206015 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002217_140 |
0.1497522013 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 14 |
Hb_000613_120 |
0.1505841844 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002997_070 |
0.1516805759 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001789_160 |
0.154541246 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 17 |
Hb_000023_370 |
0.1547352545 |
- |
- |
- |
| 18 |
Hb_031042_060 |
0.1549145466 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_021650_010 |
0.1553217461 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 20 |
Hb_001723_030 |
0.1556489869 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |