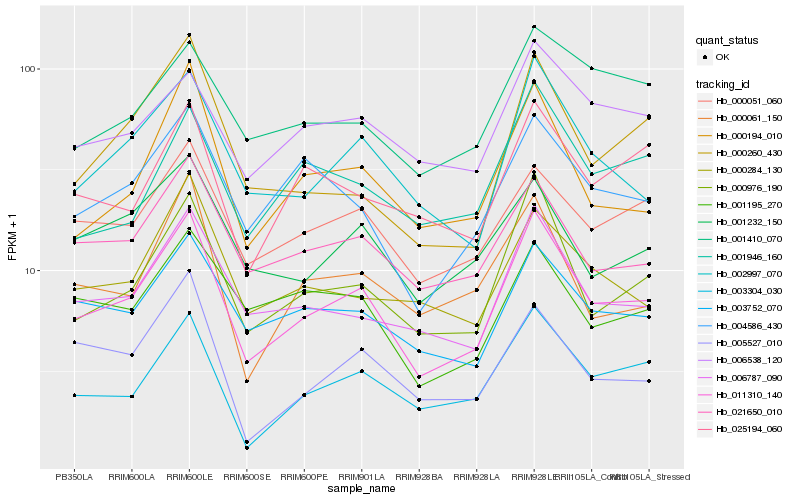
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 2 |
Hb_000976_190 |
0.0826438423 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 3 |
Hb_003304_030 |
0.0952570406 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_006787_090 |
0.0962551651 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 5 |
Hb_002997_070 |
0.0971380004 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000194_010 |
0.1037342978 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_021650_010 |
0.1056697867 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 8 |
Hb_003752_070 |
0.1064075651 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000051_060 |
0.11145396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004586_430 |
0.1118879773 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000061_150 |
0.120067008 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000284_130 |
0.1204400912 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 13 |
Hb_000260_430 |
0.1224420798 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 14 |
Hb_001232_150 |
0.1224864489 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 15 |
Hb_006538_120 |
0.1225098013 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 16 |
Hb_025194_060 |
0.1231486127 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_001946_160 |
0.1242211423 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 18 |
Hb_001195_270 |
0.1243387862 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005527_010 |
0.1260913614 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001410_070 |
0.1293119708 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |