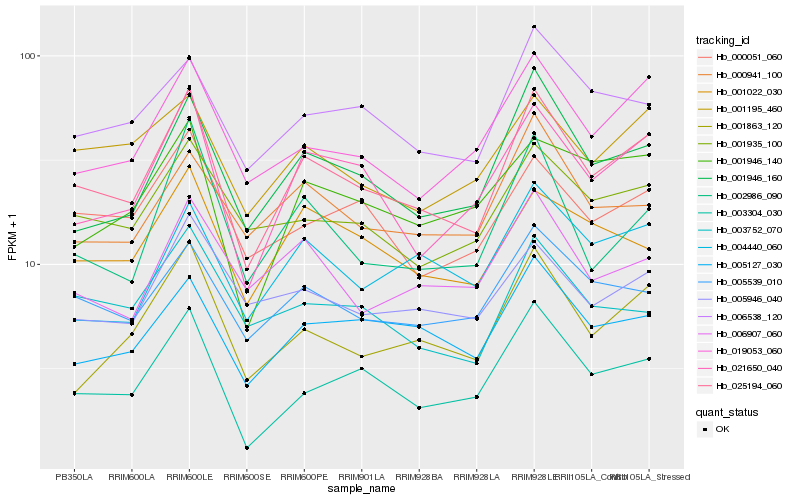
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025194_060 |
0.0 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_021650_040 |
0.0872397067 |
- |
- |
EG2771 [Manihot esculenta] |
| 3 |
Hb_001946_160 |
0.0884431781 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 4 |
Hb_003304_030 |
0.0933592394 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_019053_060 |
0.0973706548 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 6 |
Hb_005127_030 |
0.0995861997 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 7 |
Hb_005539_010 |
0.1001392167 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 8 |
Hb_001863_120 |
0.1043429894 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_006538_120 |
0.1051102983 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000051_060 |
0.1074024187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001935_100 |
0.10747203 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 12 |
Hb_002986_090 |
0.1078805149 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 13 |
Hb_001195_460 |
0.1094302969 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001022_030 |
0.1119459405 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 15 |
Hb_001946_140 |
0.1129664902 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_006907_060 |
0.1144326999 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000941_100 |
0.1147910536 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 18 |
Hb_003752_070 |
0.1153912188 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004440_060 |
0.1161227616 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 20 |
Hb_005946_040 |
0.1164445836 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |