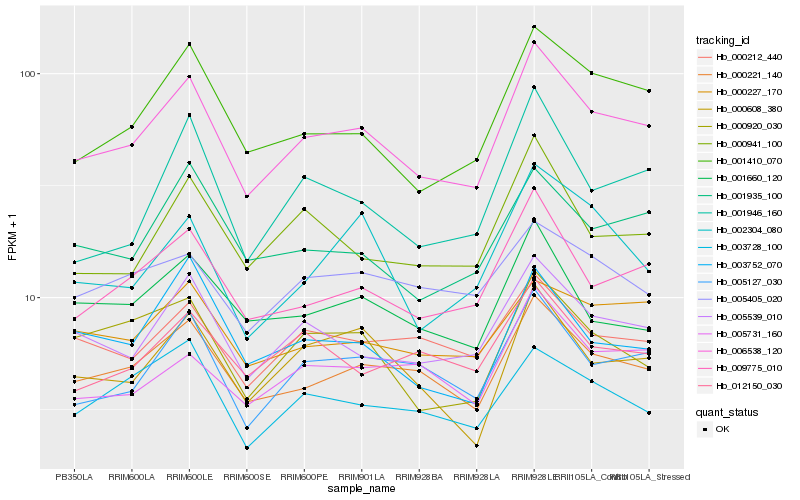
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_120 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000221_140 |
0.0617143078 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_009775_010 |
0.0689110469 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 4 |
Hb_005539_010 |
0.0710125717 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 5 |
Hb_001410_070 |
0.0733052742 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 6 |
Hb_005127_030 |
0.0746773962 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 7 |
Hb_000941_100 |
0.085388957 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 8 |
Hb_001935_100 |
0.0878488602 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 9 |
Hb_000608_380 |
0.0883989219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000212_440 |
0.0917828844 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_005731_160 |
0.0923614344 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_001660_120 |
0.0933517369 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000227_170 |
0.09361618 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_001946_160 |
0.0945625998 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 15 |
Hb_000920_030 |
0.0951601454 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_003728_100 |
0.0952691788 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012150_030 |
0.0974314726 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_005405_020 |
0.0995531959 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_003752_070 |
0.1011666579 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002304_080 |
0.1032282046 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |