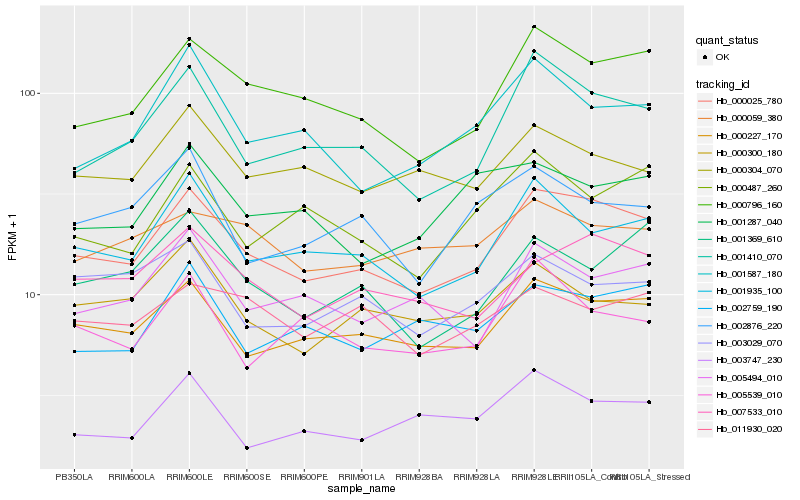
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_780 |
0.0 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 2 |
Hb_000227_170 |
0.0734608997 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 3 |
Hb_000796_160 |
0.0741040015 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001410_070 |
0.0803902051 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 5 |
Hb_001935_100 |
0.0810218802 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 6 |
Hb_007533_010 |
0.0888573504 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 7 |
Hb_005494_010 |
0.1051426414 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_001369_610 |
0.1068487988 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001587_180 |
0.1070408917 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 10 |
Hb_002759_190 |
0.1085604299 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 11 |
Hb_000487_260 |
0.1085908159 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000300_180 |
0.1106272999 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 13 |
Hb_000304_070 |
0.1110425829 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 14 |
Hb_005539_010 |
0.1122476097 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 15 |
Hb_000059_380 |
0.1123853875 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 16 |
Hb_002876_220 |
0.1129792746 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 17 |
Hb_003747_230 |
0.1130086708 |
- |
- |
PREDICTED: uncharacterized protein LOC105630239 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003029_070 |
0.1142829702 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_011930_020 |
0.116347211 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001287_040 |
0.1174056868 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |