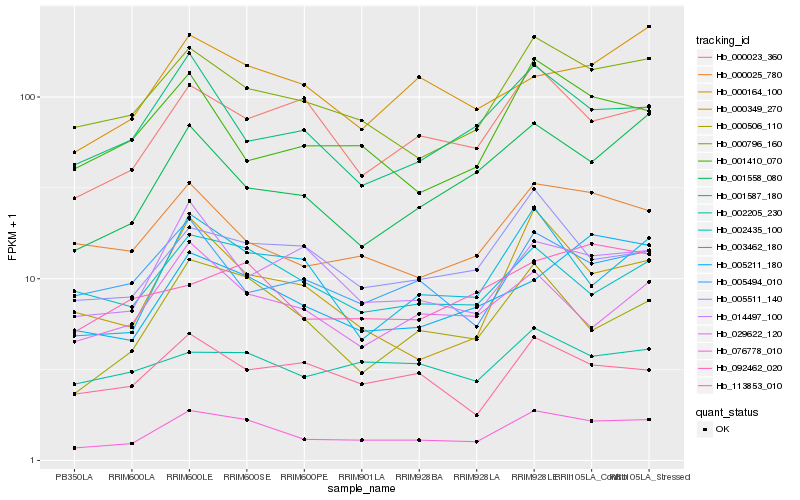
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_076778_010 |
0.0 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 2 |
Hb_000796_160 |
0.0916738896 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000506_110 |
0.1130582899 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 4 |
Hb_000025_780 |
0.1180578844 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 5 |
Hb_003462_180 |
0.1217104606 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 6 |
Hb_002435_100 |
0.1235431447 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 7 |
Hb_000023_360 |
0.1250232591 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005511_140 |
0.1265668093 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 9 |
Hb_005211_180 |
0.1266981394 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 10 |
Hb_001558_080 |
0.1280955772 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000349_270 |
0.1297737895 |
- |
- |
PREDICTED: uncharacterized protein ycf49 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000164_100 |
0.1310986003 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 13 |
Hb_029622_120 |
0.133613789 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 14 |
Hb_014497_100 |
0.1340054579 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 15 |
Hb_113853_010 |
0.1340264991 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 16 |
Hb_001587_180 |
0.134102267 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 17 |
Hb_005494_010 |
0.1348540655 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_092462_020 |
0.1350289208 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_002205_230 |
0.1360293173 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001410_070 |
0.1361855101 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |