| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
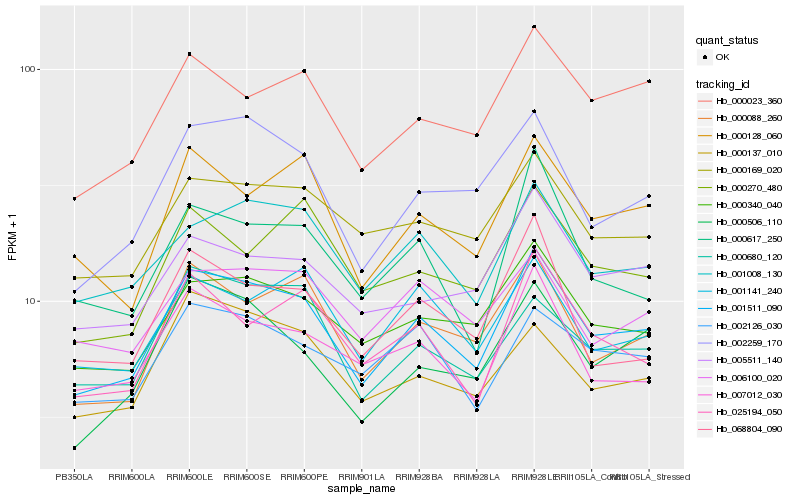
Hb_001511_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 2 |
Hb_000340_040 |
0.0766804848 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000088_260 |
0.0767051431 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 4 |
Hb_002259_170 |
0.0790026732 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001008_130 |
0.0795185541 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 6 |
Hb_000023_360 |
0.0813452279 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000169_020 |
0.0833912552 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 8 |
Hb_000128_060 |
0.0834482993 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 9 |
Hb_007012_030 |
0.0873362143 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_025194_050 |
0.0883113324 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002126_030 |
0.0905704967 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 12 |
Hb_005511_140 |
0.0906808356 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 13 |
Hb_001141_240 |
0.0915080617 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000506_110 |
0.0916116492 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 15 |
Hb_000680_120 |
0.0928574971 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 16 |
Hb_000137_010 |
0.0929173506 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 17 |
Hb_006100_020 |
0.0947730521 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 18 |
Hb_068804_090 |
0.0952678177 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 19 |
Hb_000270_480 |
0.0972017105 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 20 |
Hb_000617_250 |
0.0987284461 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |