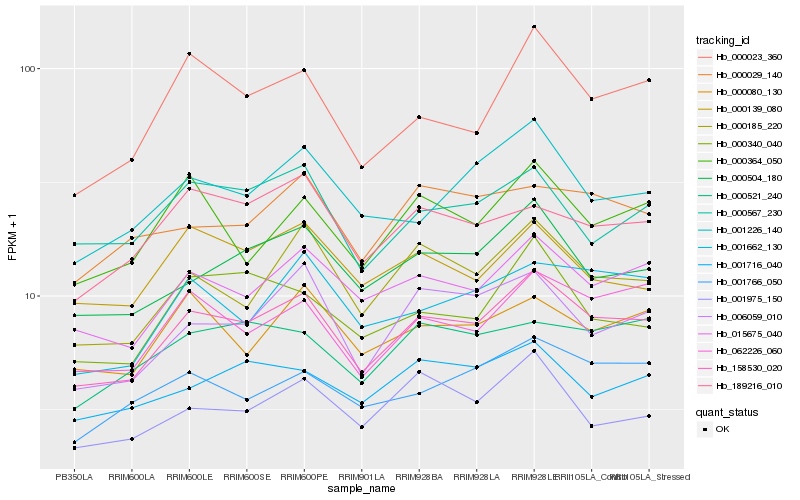
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158530_020 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 2 |
Hb_015675_040 |
0.0616252854 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 3 |
Hb_000504_180 |
0.0716894265 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 4 |
Hb_000185_220 |
0.0725578871 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 5 |
Hb_062226_060 |
0.073279461 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 6 |
Hb_006059_010 |
0.0808057019 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 7 |
Hb_000023_360 |
0.0812843687 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001662_130 |
0.0834095575 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001226_140 |
0.0836400187 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 10 |
Hb_001766_050 |
0.0842671304 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 11 |
Hb_000080_130 |
0.0857102356 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 12 |
Hb_000364_050 |
0.0861464382 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000567_230 |
0.0867502925 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 14 |
Hb_001975_150 |
0.0878465275 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 15 |
Hb_189216_010 |
0.0892130189 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001716_040 |
0.0895126081 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 17 |
Hb_000029_140 |
0.0900435915 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |
| 18 |
Hb_000340_040 |
0.0904633699 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000139_080 |
0.0907186029 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000521_240 |
0.0914670439 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |