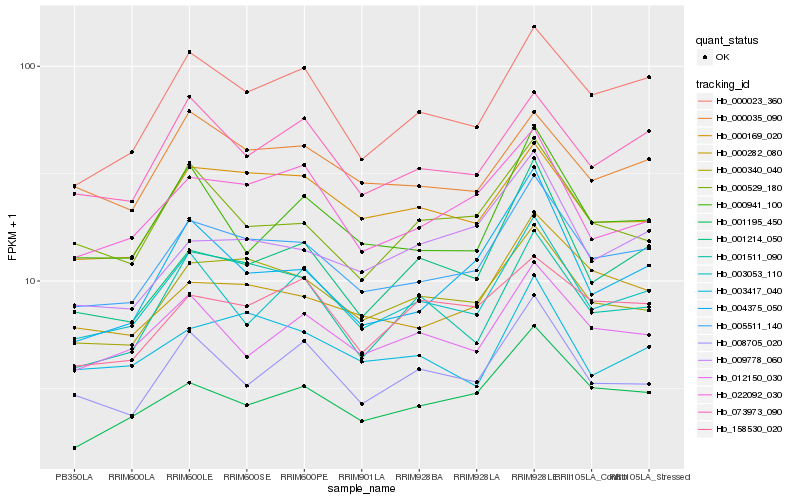
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005511_140 |
0.0 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 2 |
Hb_000340_040 |
0.063478655 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000023_360 |
0.0646167777 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000282_080 |
0.068196053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001195_450 |
0.0776457227 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 6 |
Hb_000169_020 |
0.078716576 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 7 |
Hb_073973_090 |
0.0794449227 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 8 |
Hb_022092_030 |
0.0803625039 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_003053_110 |
0.0816848621 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000035_090 |
0.0835316584 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 11 |
Hb_012150_030 |
0.0853256856 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000941_100 |
0.0874863464 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 13 |
Hb_003417_040 |
0.0898672644 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 14 |
Hb_001511_090 |
0.0906808356 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 15 |
Hb_000529_180 |
0.0907312821 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_008705_020 |
0.0910898975 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_001214_050 |
0.0915704079 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004375_050 |
0.0920970502 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_158530_020 |
0.0940082778 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 20 |
Hb_009778_060 |
0.0947925827 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |