| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
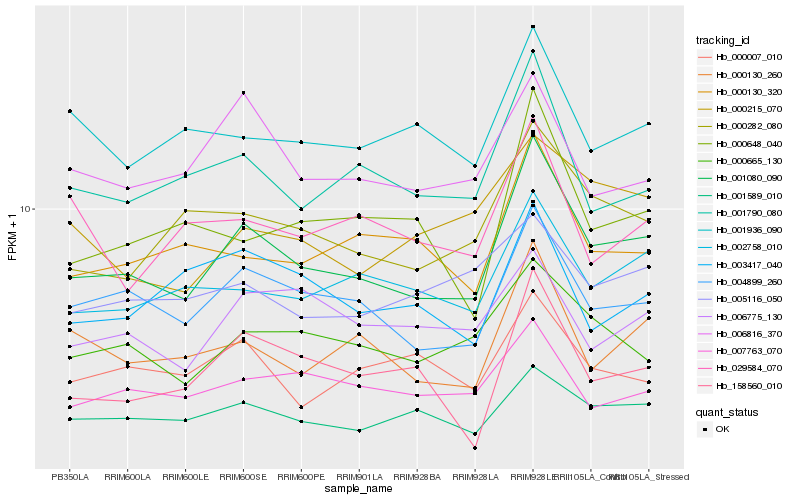
Hb_001080_090 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004899_260 |
0.0766318098 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 3 |
Hb_000282_080 |
0.0997342522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001790_080 |
0.1004696437 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_000130_260 |
0.1006056329 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001589_010 |
0.1013937113 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002758_010 |
0.1027624214 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 8 |
Hb_000665_130 |
0.1058428574 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000130_320 |
0.1093144765 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 10 |
Hb_007763_070 |
0.1135793548 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006816_370 |
0.1160462128 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 12 |
Hb_000648_040 |
0.1160900309 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_001936_090 |
0.1175685781 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_158560_010 |
0.1193160029 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 15 |
Hb_000007_010 |
0.1203694403 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 16 |
Hb_006775_130 |
0.1219780417 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 17 |
Hb_003417_040 |
0.1239725391 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 18 |
Hb_005116_050 |
0.1249763449 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000215_070 |
0.1254517585 |
- |
- |
PREDICTED: uncharacterized protein LOC105119782 isoform X1 [Populus euphratica] |
| 20 |
Hb_029584_070 |
0.1266758518 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |