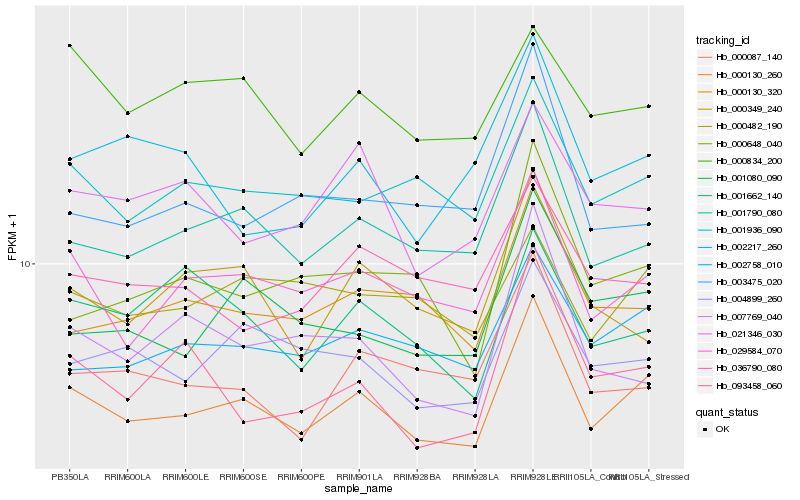
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_260 |
0.0 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 2 |
Hb_029584_070 |
0.0831815337 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 3 |
Hb_001790_080 |
0.0869456397 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 4 |
Hb_002758_010 |
0.0989132072 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 5 |
Hb_001936_090 |
0.1003387557 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001080_090 |
0.1006056329 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004899_260 |
0.106719067 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 8 |
Hb_000130_320 |
0.1084599651 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 9 |
Hb_000087_140 |
0.1159554215 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000648_040 |
0.1168146058 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_093458_060 |
0.1179815143 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 12 |
Hb_000349_240 |
0.1204123208 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_021346_030 |
0.1228684727 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 14 |
Hb_000482_190 |
0.1237909767 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 15 |
Hb_036790_080 |
0.1239461656 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 16 |
Hb_002217_260 |
0.1240071786 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 17 |
Hb_001662_140 |
0.1244777133 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007769_040 |
0.1249820363 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 19 |
Hb_003475_020 |
0.1250063868 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 20 |
Hb_000834_200 |
0.1253639849 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |