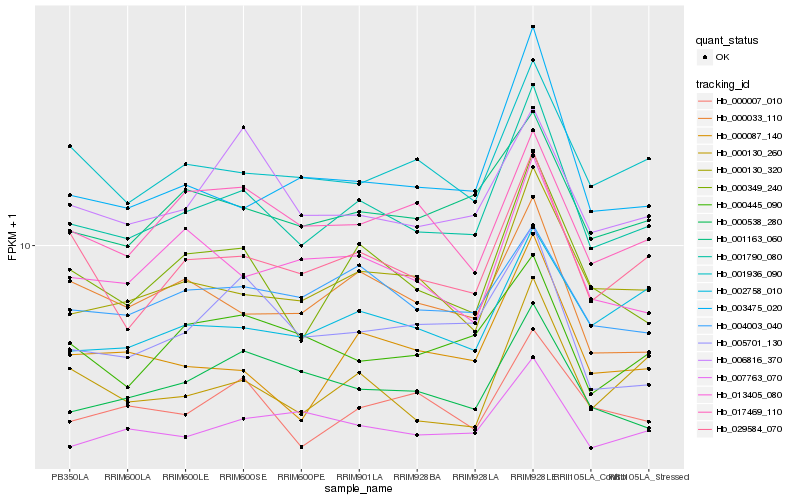
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001790_080 |
0.0 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 2 |
Hb_000130_320 |
0.0699293693 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 3 |
Hb_000349_240 |
0.0704672566 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |
| 4 |
Hb_029584_070 |
0.0763645314 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 5 |
Hb_001163_060 |
0.0772830687 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 6 |
Hb_003475_020 |
0.0817637104 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 7 |
Hb_001936_090 |
0.0841363644 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000087_140 |
0.08427958 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000130_260 |
0.0869456397 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007763_070 |
0.089160312 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000007_010 |
0.089723549 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002758_010 |
0.0911401564 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 13 |
Hb_013405_080 |
0.0914053036 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 14 |
Hb_000033_110 |
0.0914112799 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 15 |
Hb_017469_110 |
0.0937582997 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 16 |
Hb_000445_090 |
0.0947670573 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 17 |
Hb_000538_280 |
0.0959724129 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 18 |
Hb_006816_370 |
0.0961356432 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 19 |
Hb_004003_040 |
0.0961636398 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_005701_130 |
0.0962955379 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |