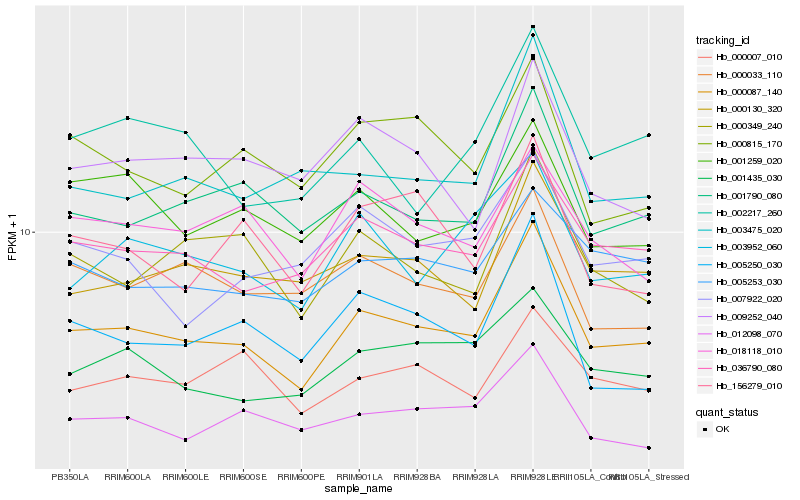
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_140 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001790_080 |
0.08427958 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 3 |
Hb_036790_080 |
0.0854670985 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 4 |
Hb_000130_320 |
0.0940936918 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 5 |
Hb_000349_240 |
0.0950946612 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001435_030 |
0.0993944766 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 7 |
Hb_003952_060 |
0.1006097253 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 8 |
Hb_003475_020 |
0.1011316412 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 9 |
Hb_005253_030 |
0.1058348217 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_005250_030 |
0.1070014021 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 11 |
Hb_002217_260 |
0.1075343009 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 12 |
Hb_000007_010 |
0.1076064867 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000033_110 |
0.1076724236 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 14 |
Hb_012098_070 |
0.107704278 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 15 |
Hb_018118_010 |
0.1079397005 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 16 |
Hb_007922_020 |
0.1095272238 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 17 |
Hb_001259_020 |
0.1096584529 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000815_170 |
0.1106774147 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 19 |
Hb_009252_040 |
0.1112765767 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_156279_010 |
0.1121285732 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |