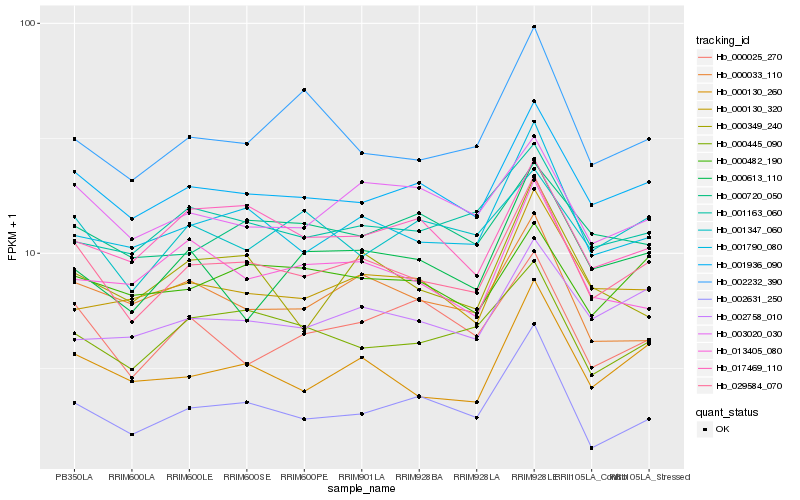
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029584_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 2 |
Hb_001936_090 |
0.0605735765 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001790_080 |
0.0763645314 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 4 |
Hb_000130_260 |
0.0831815337 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003020_030 |
0.0886370414 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_000445_090 |
0.0903872186 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 7 |
Hb_001163_060 |
0.0942040139 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 8 |
Hb_017469_110 |
0.0975002583 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 9 |
Hb_002758_010 |
0.0990210222 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 10 |
Hb_000025_270 |
0.1001816745 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_000482_190 |
0.1002228948 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 12 |
Hb_000033_110 |
0.1008182547 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 13 |
Hb_000130_320 |
0.1012452496 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 14 |
Hb_002232_390 |
0.1018068225 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 15 |
Hb_002631_250 |
0.1027491002 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 16 |
Hb_000720_050 |
0.1031381564 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001347_060 |
0.1050482786 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 18 |
Hb_000613_110 |
0.105323632 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 19 |
Hb_013405_080 |
0.1064028009 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 20 |
Hb_000349_240 |
0.1070524428 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |