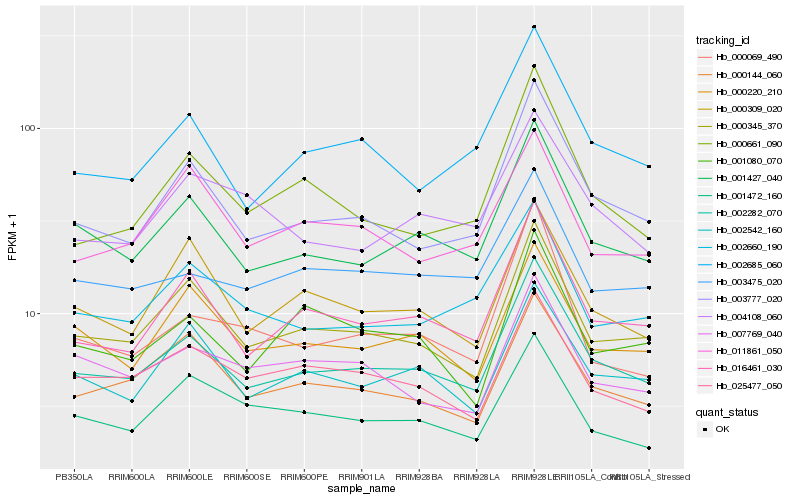
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002282_070 |
0.0 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 2 |
Hb_001427_040 |
0.0577864004 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 3 |
Hb_016461_030 |
0.0612862576 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000345_370 |
0.0640764716 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 5 |
Hb_003777_020 |
0.0657131302 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 6 |
Hb_002685_060 |
0.0805126532 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 7 |
Hb_000144_060 |
0.0853293367 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002542_160 |
0.0855619595 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003475_020 |
0.0881868221 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 10 |
Hb_000220_210 |
0.0891918647 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000309_020 |
0.0929289113 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 12 |
Hb_002660_190 |
0.0929488231 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 13 |
Hb_025477_050 |
0.0951400558 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_000661_090 |
0.0959971643 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007769_040 |
0.1013424811 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_004108_060 |
0.1024286557 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 17 |
Hb_001080_070 |
0.1042836537 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 18 |
Hb_001472_160 |
0.1064101044 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 19 |
Hb_000069_490 |
0.1080181524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011861_050 |
0.10842205 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |