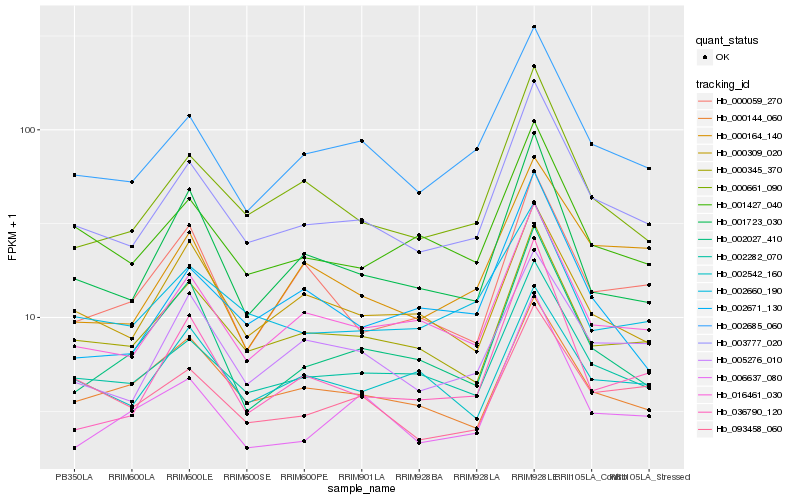
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003777_020 |
0.0 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 2 |
Hb_002282_070 |
0.0657131302 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 3 |
Hb_002685_060 |
0.0675093465 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 4 |
Hb_016461_030 |
0.0745819103 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000661_090 |
0.0789102405 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000345_370 |
0.0823556093 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 7 |
Hb_001427_040 |
0.0894520826 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 8 |
Hb_005276_010 |
0.0922023502 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 9 |
Hb_001723_030 |
0.0983462111 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002660_190 |
0.1035109044 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 11 |
Hb_093458_060 |
0.1056082301 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 12 |
Hb_000144_060 |
0.1066088215 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_006637_080 |
0.107604947 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000309_020 |
0.1079111636 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 15 |
Hb_000164_140 |
0.1092759065 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_002027_410 |
0.109329768 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002542_160 |
0.111918039 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000059_270 |
0.1140476493 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 19 |
Hb_002671_130 |
0.1152402054 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 20 |
Hb_036790_120 |
0.1159754677 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |