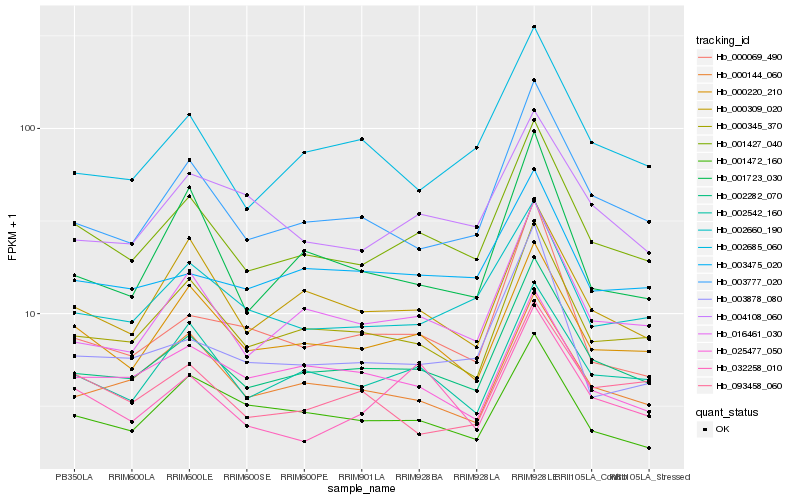
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 2 |
Hb_002282_070 |
0.0577864004 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 3 |
Hb_000220_210 |
0.0769550167 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 4 |
Hb_016461_030 |
0.0813955613 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_002542_160 |
0.0829712649 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000345_370 |
0.0836939789 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 7 |
Hb_002660_190 |
0.0864093873 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 8 |
Hb_003777_020 |
0.0894520826 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 9 |
Hb_000309_020 |
0.0958747488 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 10 |
Hb_032258_010 |
0.1004341848 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 11 |
Hb_003475_020 |
0.1062745143 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 12 |
Hb_000144_060 |
0.1067600622 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_004108_060 |
0.1074499392 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 14 |
Hb_001472_160 |
0.1097102093 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_002685_060 |
0.1100203399 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 16 |
Hb_003878_080 |
0.1160381306 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 17 |
Hb_025477_050 |
0.1162563822 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_001723_030 |
0.1168460985 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000069_490 |
0.1176874784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_093458_060 |
0.1195932434 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |