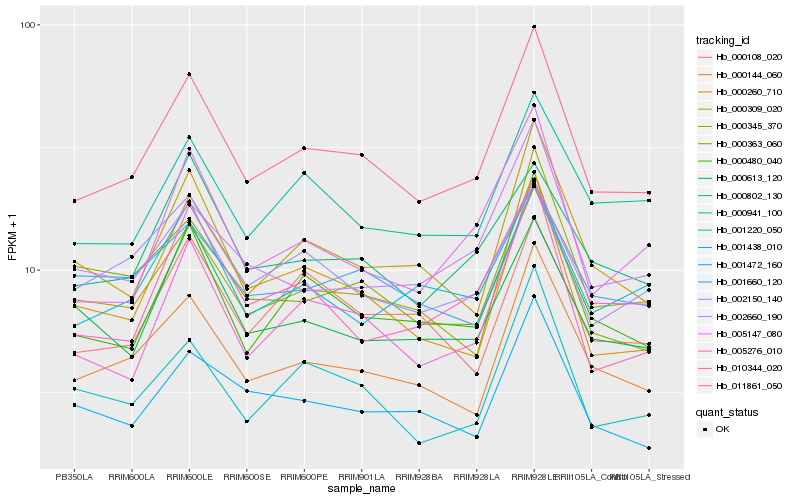
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011861_050 |
0.0 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_000480_040 |
0.0669674321 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 3 |
Hb_000144_060 |
0.0714021622 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000345_370 |
0.0819135493 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 5 |
Hb_000309_020 |
0.0819463521 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 6 |
Hb_000941_100 |
0.0862424943 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 7 |
Hb_005147_080 |
0.0880282286 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000260_710 |
0.0897785895 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001438_010 |
0.0912530939 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000802_130 |
0.0914284198 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 11 |
Hb_001472_160 |
0.0928777269 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 12 |
Hb_010344_020 |
0.0934382352 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001220_050 |
0.0943401524 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 14 |
Hb_002660_190 |
0.0946872565 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 15 |
Hb_002150_140 |
0.0961507435 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 16 |
Hb_005276_010 |
0.0974721963 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 17 |
Hb_000613_120 |
0.0980408377 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000108_020 |
0.0984544101 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 19 |
Hb_001660_120 |
0.0992343005 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000363_060 |
0.0997029161 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |