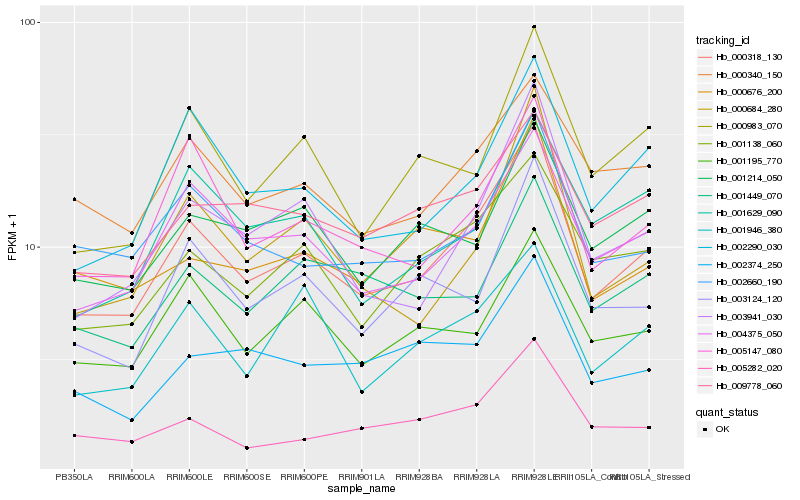
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_130 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_004375_050 |
0.0891305603 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_003941_030 |
0.0998487919 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 4 |
Hb_001138_060 |
0.1002615305 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 5 |
Hb_005147_080 |
0.1023308518 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_002290_030 |
0.1033146756 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002660_190 |
0.1099187956 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 8 |
Hb_003124_120 |
0.1113039733 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 9 |
Hb_000340_150 |
0.1114047957 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 10 |
Hb_000983_070 |
0.1128762348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005282_020 |
0.1141300669 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 12 |
Hb_000676_200 |
0.1147258586 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 13 |
Hb_009778_060 |
0.1148596532 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 14 |
Hb_001195_770 |
0.1201112186 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 15 |
Hb_002374_250 |
0.1201178368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001214_050 |
0.1208578736 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001946_380 |
0.1212296096 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 18 |
Hb_001449_070 |
0.1218060718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000684_280 |
0.1218568984 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001629_090 |
0.1250127213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |