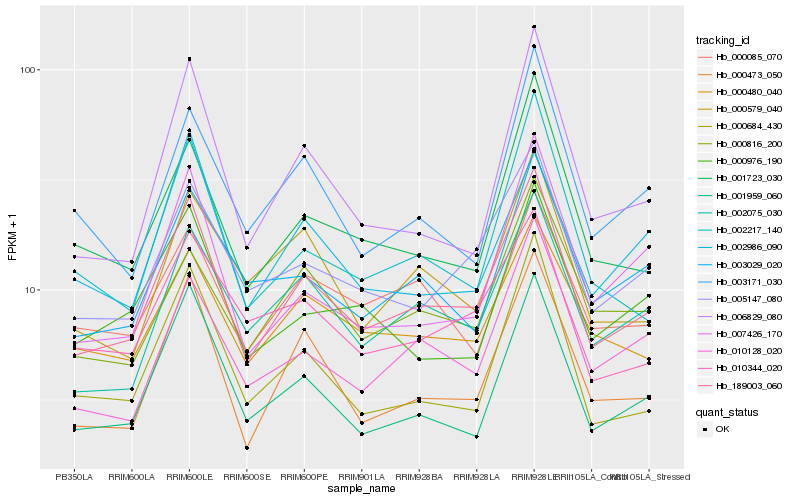
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189003_060 |
0.0 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000473_050 |
0.0851732977 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 3 |
Hb_005147_080 |
0.0881547474 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_002075_030 |
0.0883170354 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001959_060 |
0.0895357521 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000684_430 |
0.0997266601 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000480_040 |
0.0999248511 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 8 |
Hb_001723_030 |
0.1005542132 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_010128_020 |
0.102104646 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 10 |
Hb_006829_080 |
0.1026656571 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 11 |
Hb_010344_020 |
0.1032501509 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003029_020 |
0.1062598992 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 13 |
Hb_002217_140 |
0.1075038783 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 14 |
Hb_000579_040 |
0.1087054187 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 15 |
Hb_007426_170 |
0.1090680564 |
transcription factor |
TF Family: TCP |
- |
| 16 |
Hb_000816_200 |
0.1096023313 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 17 |
Hb_000085_070 |
0.1113464979 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_003171_030 |
0.1128872313 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000976_190 |
0.113253218 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 20 |
Hb_002986_090 |
0.1137646598 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |