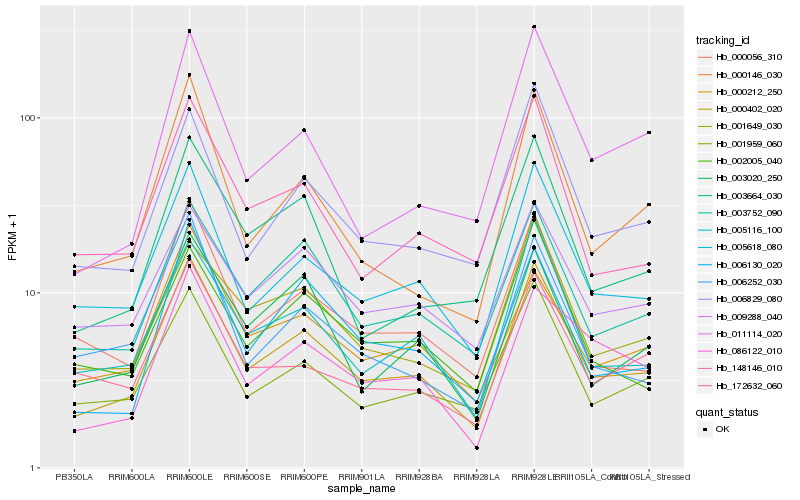
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_020 |
0.0 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_005618_080 |
0.0751996034 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005116_100 |
0.0900626947 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 4 |
Hb_001959_060 |
0.0935072173 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003752_090 |
0.0948105879 |
- |
- |
chitinase, putative [Ricinus communis] |
| 6 |
Hb_006829_080 |
0.1005389553 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 7 |
Hb_000056_310 |
0.1019973807 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 8 |
Hb_148146_010 |
0.1039004041 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 9 |
Hb_001649_030 |
0.1049759171 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000146_030 |
0.1052904624 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006130_020 |
0.1070981988 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000212_250 |
0.1091075282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009288_040 |
0.1099707151 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 14 |
Hb_003664_030 |
0.1121344057 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 15 |
Hb_003020_250 |
0.1132100893 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 16 |
Hb_086122_010 |
0.1138240024 |
- |
- |
PREDICTED: immunoglobulin-like domain-containing receptor 1 [Jatropha curcas] |
| 17 |
Hb_006252_030 |
0.1146822295 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 18 |
Hb_002005_040 |
0.1148540219 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_172632_060 |
0.1152706613 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011114_020 |
0.1155249903 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |