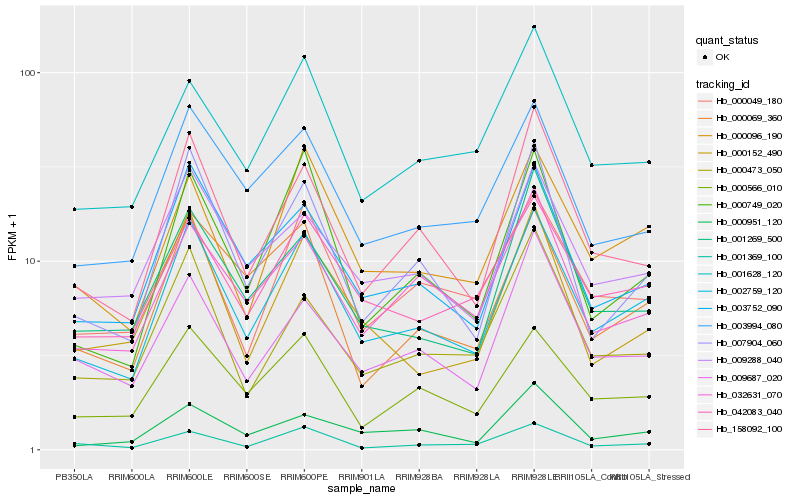
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003752_090 |
0.0882570361 |
- |
- |
chitinase, putative [Ricinus communis] |
| 3 |
Hb_000096_190 |
0.0931515463 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_000566_010 |
0.095761533 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 5 |
Hb_042083_040 |
0.0969749846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_158092_100 |
0.0999981201 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 7 |
Hb_003994_080 |
0.1033532872 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 8 |
Hb_007904_060 |
0.1034212973 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 9 |
Hb_000749_020 |
0.1063079647 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_009288_040 |
0.1075860566 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 11 |
Hb_000049_180 |
0.109505619 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 12 |
Hb_000069_360 |
0.113173041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001628_120 |
0.1137956395 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 14 |
Hb_001269_500 |
0.1152820057 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 15 |
Hb_032631_070 |
0.1154865287 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000152_490 |
0.1175499921 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 17 |
Hb_000473_050 |
0.1193637591 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 18 |
Hb_009687_020 |
0.1200957756 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 19 |
Hb_000951_120 |
0.1206711359 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 20 |
Hb_001369_100 |
0.1213651599 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |