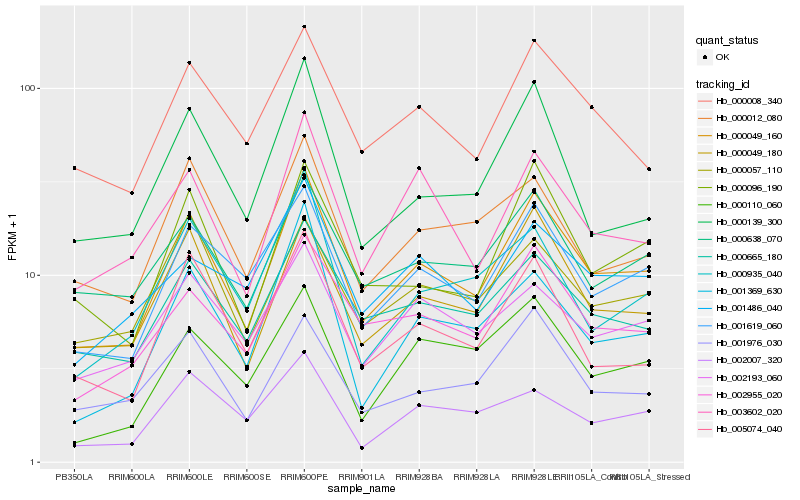
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_160 |
0.0 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 2 |
Hb_001619_060 |
0.0880694231 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000049_180 |
0.0883389881 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 4 |
Hb_002955_020 |
0.0960798215 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000096_190 |
0.0972675614 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_000665_180 |
0.0979261919 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_000008_340 |
0.1023271168 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 8 |
Hb_003602_020 |
0.1068714878 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_005074_040 |
0.1073506682 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000638_070 |
0.1075005335 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 11 |
Hb_001976_030 |
0.1081802319 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000012_080 |
0.1118404488 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 13 |
Hb_000139_300 |
0.1119076931 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 14 |
Hb_002193_060 |
0.1151824275 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 15 |
Hb_000057_110 |
0.1170630744 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 16 |
Hb_000110_060 |
0.1185905548 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 17 |
Hb_001486_040 |
0.1203577132 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000935_040 |
0.1204166246 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_001369_630 |
0.1221966622 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 20 |
Hb_002007_320 |
0.1236860753 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |