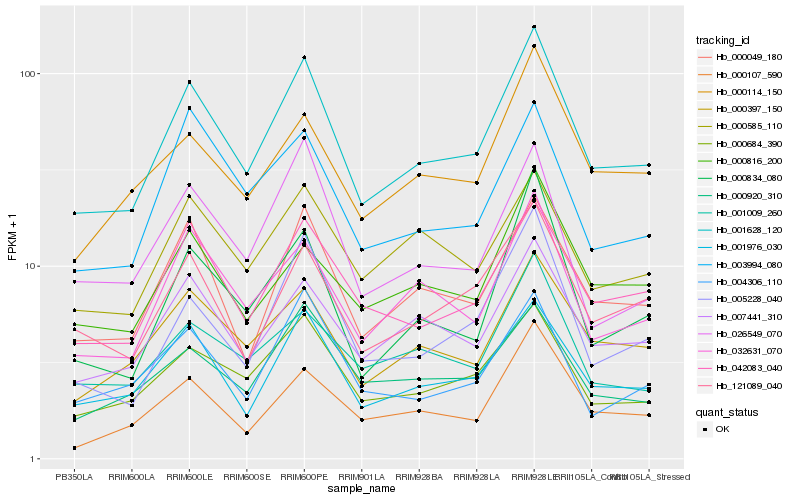
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001628_120 |
0.0 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 2 |
Hb_001976_030 |
0.0792784707 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000684_390 |
0.0847097709 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 4 |
Hb_042083_040 |
0.0871008195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000049_180 |
0.0871427482 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 6 |
Hb_007441_310 |
0.0884140342 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000397_150 |
0.0911428273 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 8 |
Hb_032631_070 |
0.0911460588 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_121089_040 |
0.0950729406 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 10 |
Hb_005228_040 |
0.1033893358 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 11 |
Hb_000920_310 |
0.1053011035 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_000107_590 |
0.1061931624 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 13 |
Hb_000834_080 |
0.1064273803 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 14 |
Hb_000114_150 |
0.1066369068 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 15 |
Hb_003994_080 |
0.1099788808 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 16 |
Hb_000585_110 |
0.1106389571 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 17 |
Hb_004306_110 |
0.1106508016 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 18 |
Hb_001009_260 |
0.1115463263 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000816_200 |
0.1115879382 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 20 |
Hb_026549_070 |
0.1133491068 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |