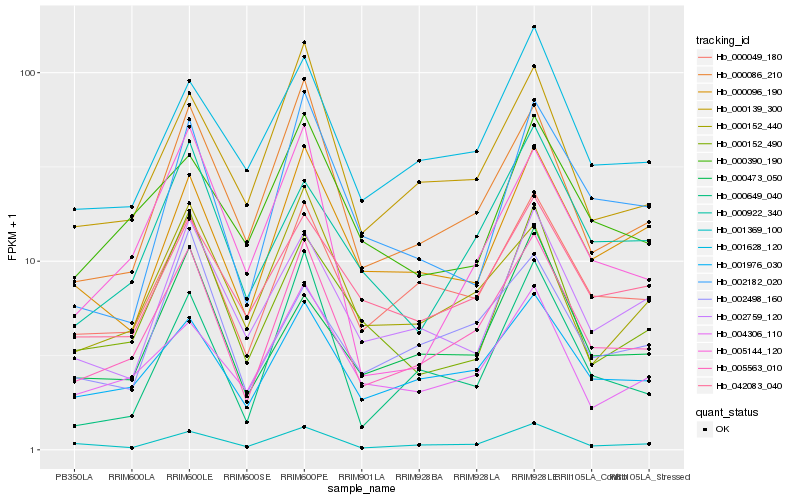
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005563_010 |
0.0 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 2 |
Hb_001976_030 |
0.0908330588 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000049_180 |
0.0951468879 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 4 |
Hb_000086_210 |
0.0998689694 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_001369_100 |
0.110861338 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 6 |
Hb_000139_300 |
0.1166803267 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 7 |
Hb_001628_120 |
0.12419205 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 8 |
Hb_000096_190 |
0.1286606685 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_004306_110 |
0.1298496817 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 10 |
Hb_042083_040 |
0.1324086025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000922_340 |
0.132932008 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 12 |
Hb_000152_440 |
0.1336371742 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 13 |
Hb_002759_120 |
0.1340938589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000473_050 |
0.1367265818 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 15 |
Hb_000152_490 |
0.1378594152 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 16 |
Hb_005144_120 |
0.1414602794 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 17 |
Hb_000649_040 |
0.1415438953 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 18 |
Hb_002182_020 |
0.1423170573 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000390_190 |
0.1433677628 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_002498_160 |
0.1434996666 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |