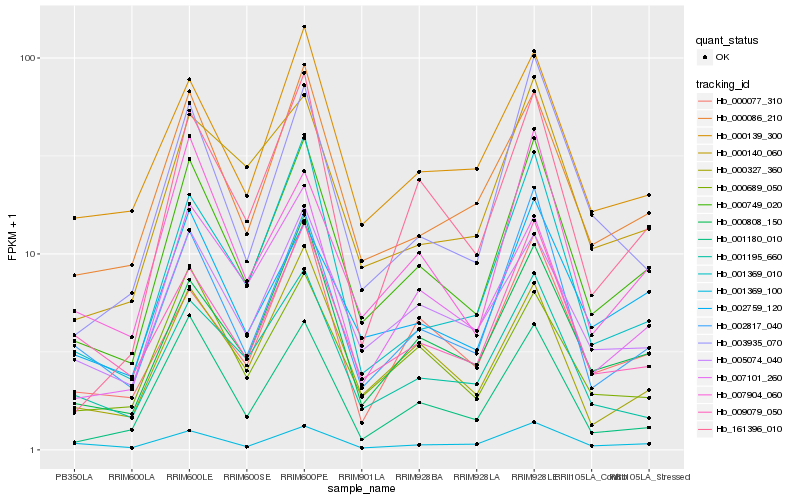
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_020 |
0.0 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_002817_040 |
0.0936770539 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 3 |
Hb_000086_210 |
0.0963287115 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_000808_150 |
0.0995873312 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 5 |
Hb_002759_120 |
0.1063079647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005074_040 |
0.1099500405 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_007904_060 |
0.1128884256 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 8 |
Hb_000139_300 |
0.1131908777 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 9 |
Hb_000327_360 |
0.1136679584 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_001369_010 |
0.1136998504 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 11 |
Hb_007101_260 |
0.1150510745 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 12 |
Hb_000140_060 |
0.1151492095 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 13 |
Hb_161396_010 |
0.1159220095 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 14 |
Hb_001195_660 |
0.1178232071 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 15 |
Hb_003935_070 |
0.1182261433 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001369_100 |
0.1196013821 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 17 |
Hb_000689_050 |
0.119647975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009079_050 |
0.122700804 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 19 |
Hb_000077_310 |
0.1228510681 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 20 |
Hb_001180_010 |
0.1249559218 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |